Transient diffusion - partial differential equations
Posted April 02, 2013 at 09:27 AM | categories: pde | tags: mass transfer
Updated April 02, 2013 at 09:44 AM
We want to solve for the concentration profile of component that diffuses into a 1D rod, with an impermeable barrier at the end. The PDE governing this situation is:
\(\frac{\partial C}{\partial t} = D \frac{\partial^2 C}{\partial x^2}\)
at \(t=0\), in this example we have \(C_0(x) = 0\) as an initial condition, with boundary conditions \(C(0,t)=0.1\) and \(\partial C/ \partial x(L,t)=0\).
We are going to discretize this equation in both time and space to arrive at the solution. We will let \(i\) be the index for the spatial discretization, and \(j\) be the index for the temporal discretization. The discretization looks like this.
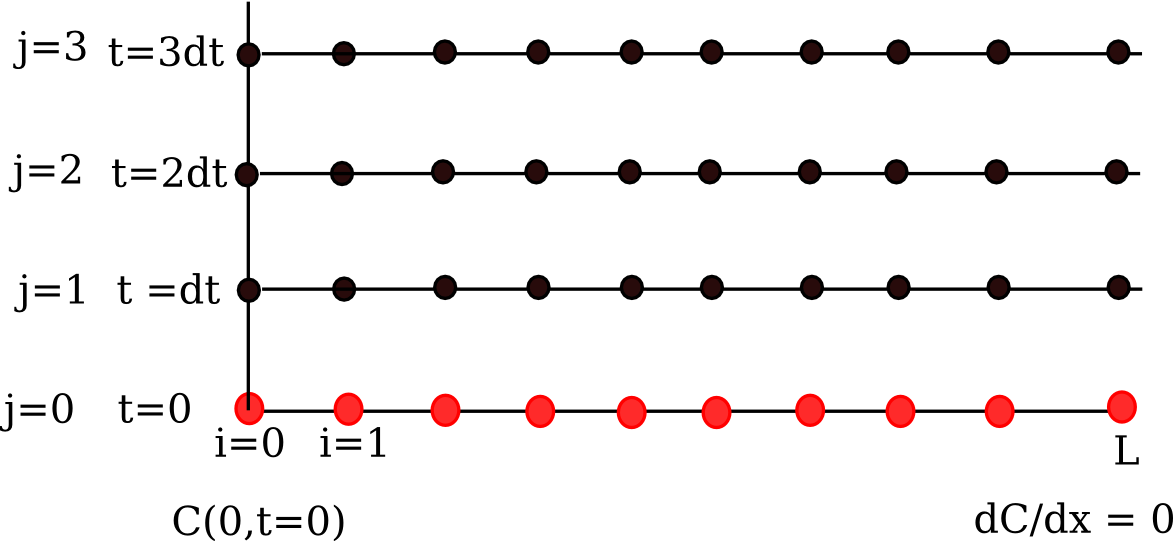
Note that we cannot use the method of lines as we did before because we have the derivative-based boundary condition at one of the boundaries.
We approximate the time derivative as:
\(\frac{\partial C}{\partial t} \bigg| _{i,j} \approx \frac{C_{i,j+1} - C_{i,j}}{\Delta t} \)
\(\frac{\partial^2 C}{\partial x^2} \bigg| _{i,j} \approx \frac{C_{i+1,j} - 2 C_{i,j} + C_{i-1,j}}{h^2} \)
We define \(\alpha = \frac{D \Delta t}{h^2}\), and from these two approximations and the PDE, we solve for the unknown solution at a later time step as:
\(C_{i, j+1} = \alpha C_{i+1,j} + (1 - 2 \alpha) C_{i,j} + \alpha C_{i-1,j} \)
We know \(C_{i,j=0}\) from the initial conditions, so we simply need to iterate to evaluate \(C_{i,j}\), which is the solution at each time step.
See also: http://www3.nd.edu/~jjwteach/441/PdfNotes/lecture16.pdf
import numpy as np import matplotlib.pyplot as plt N = 20 # number of points to discretize L = 1.0 X = np.linspace(0, L, N) # position along the rod h = L / (N - 1) # discretization spacing C0t = 0.1 # concentration at x = 0 D = 0.02 tfinal = 50.0 Ntsteps = 1000 dt = tfinal / (Ntsteps - 1) t = np.linspace(0, tfinal, Ntsteps) alpha = D * dt / h**2 print alpha C_xt = [] # container for all the time steps # initial condition at t = 0 C = np.zeros(X.shape) C[0] = C0t C_xt += [C] for j in range(1, Ntsteps): N = np.zeros(C.shape) N[0] = C0t N[1:-1] = alpha*C[2:] + (1 - 2 * alpha) * C[1:-1] + alpha * C[0:-2] N[-1] = N[-2] # derivative boundary condition flux = 0 C[:] = N C_xt += [N] # plot selective solutions if j in [1,2,5,10,20,50,100,200,500]: plt.plot(X, N, label='t={0:1.2f}'.format(t[j])) plt.xlabel('Position in rod') plt.ylabel('Concentration') plt.title('Concentration at different times') plt.legend(loc='best') plt.savefig('images/transient-diffusion-temporal-dependence.png') C_xt = np.array(C_xt) plt.figure() plt.plot(t, C_xt[:,5], label='x={0:1.2f}'.format(X[5])) plt.plot(t, C_xt[:,10], label='x={0:1.2f}'.format(X[10])) plt.plot(t, C_xt[:,15], label='x={0:1.2f}'.format(X[15])) plt.plot(t, C_xt[:,19], label='x={0:1.2f}'.format(X[19])) plt.legend(loc='best') plt.xlabel('Time') plt.ylabel('Concentration') plt.savefig('images/transient-diffusion-position-dependence.png') plt.show()
0.361361361361
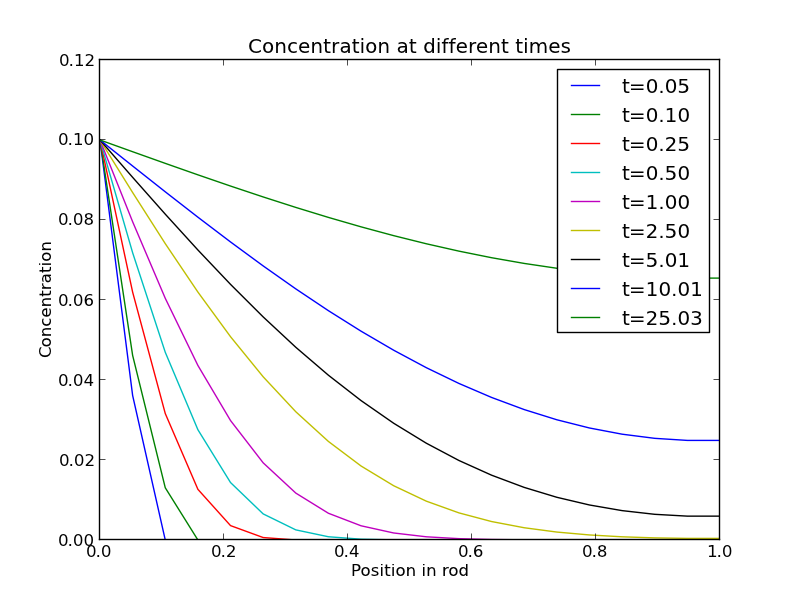
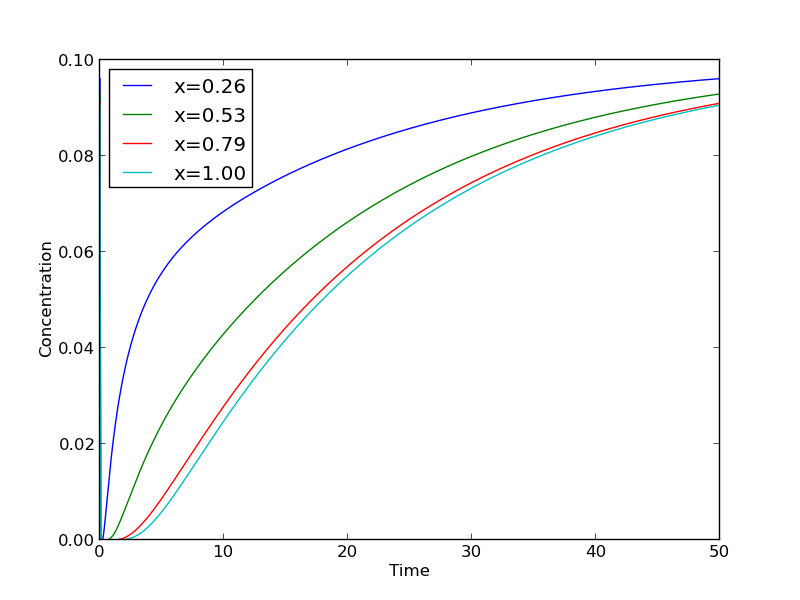
The solution is somewhat sensitive to the choices of time step and spatial discretization. If you make the time step too big, the method is not stable, and large oscillations may occur.
Copyright (C) 2013 by John Kitchin. See the License for information about copying.