Units in ODEs
Posted March 25, 2013 at 09:58 AM | categories: odes, units | tags:
We reconsider a simple ODE but this time with units. We will use the quantities package again.
Here is the ODE, \(\frac{dCa}{dt} = -k Ca\) with \(C_A(0) = 1.0\) mol/L and \(k = 0.23\) 1/s. Compute the concentration after 5 s.
import quantities as u k = 0.23 / u.s Ca0 = 1 * u.mol / u.L def dCadt(Ca, t): return -k * Ca import numpy as np from scipy.integrate import odeint tspan = np.linspace(0, 5) * u.s sol = odeint(dCadt, Ca0, tspan) print sol[-1]
[ 0.31663678]
No surprise, the units are lost. Now we start wrapping odeint. We wrap everything, and then test two examples including a single ODE, and a coupled set of ODEs with mixed units.
import quantities as u import matplotlib.pyplot as plt import numpy as np from scipy.integrate import odeint as _odeint def odeint(func, y0, t, args=(), Dfun=None, col_deriv=0, full_output=0, ml=None, mu=None, rtol=None, atol=None, tcrit=None, h0=0.0, hmax=0.0, hmin=0.0, ixpr=0, mxstep=0, mxhnil=0, mxordn=12, mxords=5, printmessg=0): def wrapped_func(Y0, T, *args): # put units on T if they are on the original t # check for units so we don't put them on twice if not hasattr(T, 'units') and hasattr(t, 'units'): T = T * t.units # now for the dependent variable units. Y0 may be a scalar or # a list or an array. we want to check each element of y0 for # units, and add them to the corresponding element of Y0 if we # need to. try: uY0 = [x for x in Y0] # a list copy of contents of Y0 # this works if y0 is an iterable, eg. a list or array for i, yi in enumerate(y0): if not hasattr(uY0[i],'units') and hasattr(yi, 'units'): uY0[i] = uY0[i] * yi.units except TypeError: # we have a scalar if not hasattr(Y0, 'units') and hasattr(y0, 'units'): uY0 = Y0 * y0.units val = func(uY0, t, *args) try: return np.array([float(x) for x in val]) except TypeError: return float(val) if full_output: y, infodict = _odeint(wrapped_func, y0, t, args, Dfun, col_deriv, full_output, ml, mu, rtol, atol, tcrit, h0, hmax, hmin, ixpr, mxstep, mxhnil, mxordn, mxords, printmessg) else: y = _odeint(wrapped_func, y0, t, args, Dfun, col_deriv, full_output, ml, mu, rtol, atol, tcrit, h0, hmax, hmin, ixpr, mxstep, mxhnil, mxordn, mxords, printmessg) # now we need to put units onto the solution units should be the # same as y0. We cannot put mixed units in an array, so, we return a list m,n = y.shape # y is an ndarray, so it has a shape if n > 1: # more than one equation, we need a list uY = [0 for yi in range(n)] for i, yi in enumerate(y0): if not hasattr(uY[i],'units') and hasattr(yi, 'units'): uY[i] = y[:,i] * yi.units else: uY[i] = y[:,i] else: uY = y * y0.units y = uY if full_output: return y, infodict else: return y ################################################################## # test a single ODE k = 0.23 / u.s Ca0 = 1 * u.mol / u.L def dCadt(Ca, t): return -k * Ca tspan = np.linspace(0, 5) * u.s sol = odeint(dCadt, Ca0, tspan) print sol[-1] plt.plot(tspan, sol) plt.xlabel('Time ({0})'.format(tspan.dimensionality.latex)) plt.ylabel('$C_A$ ({0})'.format(sol.dimensionality.latex)) plt.savefig('images/ode-units-ca.png') ################################################################## # test coupled ODEs lbmol = 453.59237*u.mol kprime = 0.0266 * lbmol / u.hr / u.lb Fa0 = 1.08 * lbmol / u.hr alpha = 0.0166 / u.lb epsilon = -0.15 def dFdW(F, W, alpha0): X, y = F dXdW = kprime / Fa0 * (1.0 - X)/(1.0 + epsilon * X) * y dydW = - alpha0 * (1.0 + epsilon * X) / (2.0 * y) return [dXdW, dydW] X0 = 0.0 * u.dimensionless y0 = 1.0 # initial conditions F0 = [X0, y0] # one without units, one with units, both are dimensionless wspan = np.linspace(0,60) * u.lb sol = odeint(dFdW, F0, wspan, args=(alpha,)) X, y = sol print 'Test 2' print X[-1] print y[-1] plt.figure() plt.plot(wspan, X, wspan, y) plt.legend(['X','$P/P_0$']) plt.xlabel('Catalyst weight ({0})'.format(wspan.dimensionality.latex)) plt.savefig('images/ode-coupled-units-pdrpo.png')
[ 0.31663678] mol/L Test 2 0.665569578156 dimensionless 0.263300470681
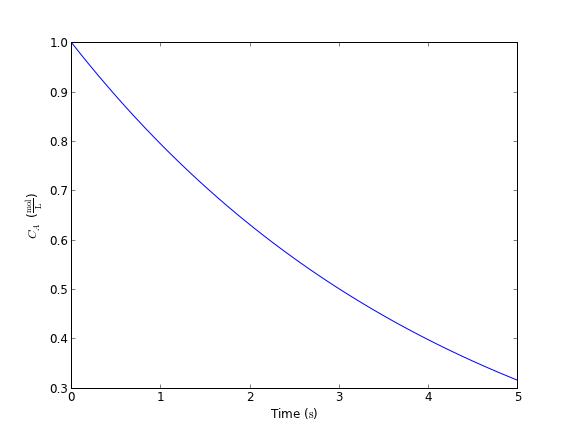
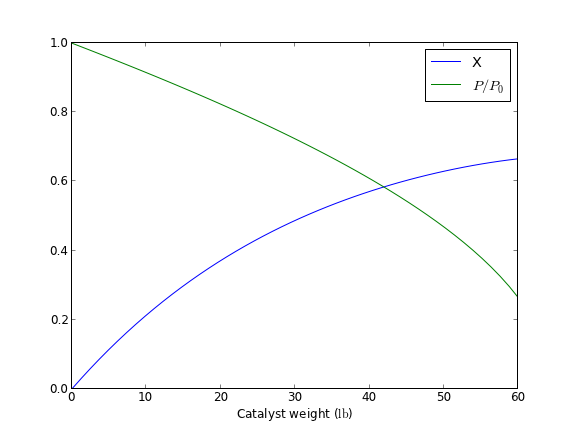
That is not too bad. This is another example of a function you would want to save in a module for reuse. There is one bad feature of the wrapped odeint function, and that is that it changes the solution for coupled ODEs from an ndarray to a list. That is necessary because you apparently cannot have mixed units in an ndarray. It is fine, however, to have a list of mixed units. This is not a huge problem, but it changes the syntax for plotting results for the wrapped odeint function compared to the unwrapped function without units.
Copyright (C) 2013 by John Kitchin. See the License for information about copying.