One of my students asked about highlighting text in emacs for note-taking. I can see some advantages for doing it while teaching, for example, and for students studying, so here we we work it out.
You will definitely want to see the video on this one, the highlights do not show up in the published html. https://www.youtube.com/watch?v=Cvz2tiT12-I
For temporary use, highlighting is pretty easy, you just set a property on a region, e.g. the background color. For example, we can do this:
; this seems to be necessary to get the tooltips to work.
(setq font-lock-extra-managed-props (delq 'help-echo font-lock-extra-managed-props))
(defun highlight-region (beg end)
(interactive "r")
(set-text-properties
beg end
'(font-lock-face (:background "Light Salmon")
highlighted t
help-echo "highlighted")))
(global-set-key (kbd "s-h") 'highlight-region)
highlight-region
This sets the background color, and another property "highlighted" that we will use later. The trouble is this is transient. When I close the file, the highlights are lost. We can save them to a file though, and reload them later. As long as we are diligent about that we should be able to provide persistent highlights.
First we need a function to get all the highlights, their start and end, their color, and if there is a help-echo which provides a tooltip. We will see why later. Here we loop through the buffer collecting highlights, and return a list of them.
(defun highlight-get-highlights ()
"Scan buffer for list of highlighted regions.
These are defined only by the highlighted property. That means
adjacent highlighted regions will be merged into one region with
the color of the first one."
(save-excursion
(goto-char (point-min))
(let ((highlights '())
(p)
(beg)
(end)
(note)
(color))
;; corner case of first point being highlighted
(when (get-text-property (point) 'highlighted)
(setq beg (point)
end (next-single-property-change (point) 'highlighted)
color (background-color-at-point)
help-echo (get-text-property (point) 'help-echo))
(add-to-list 'highlights (list beg end color help-echo) t)
(goto-char end))
;; Now the rest of the buffer
(while (setq p (next-single-property-change (point) 'highlighted))
(setq beg (goto-char p))
(setq color (background-color-at-point))
(setq note (get-text-property (point) 'help-echo))
(setq end (next-single-property-change (point) 'highlighted))
(when (and beg end)
(goto-char end)
(add-to-list 'highlights (list beg
end
color
note)
t)
(goto-char end)))
highlights)))
(highlight-get-highlights)
| 438 |
454 |
Light Salmon |
highlighted |
| 1014 |
1031 |
Light Salmon |
highlighted |
Next, we generate a filename, and a function to save the highlights to disk. We make it a hook function that runs every time we save the buffer.
(defun highlight-save-filename ()
"Return name of file to save overlays in."
(when (buffer-file-name)
(concat "." (file-name-nondirectory (buffer-file-name)) ".highlights")))
(defun highlight-save ()
"Loop through buffer and save regions with property highlighted.
Save beginning, end of each region, color and help-echo on the
first character of the region. Delete highlight file if it is empty."
(interactive)
(let ((fname (highlight-save-filename))
(highlights (highlight-get-highlights)))
(if (and fname highlights)
(with-temp-file fname
(print highlights (current-buffer)))
;; get rid of file if there are not highlights
(when (and fname (file-exists-p fname))
(delete-file fname)))))
(add-hook 'after-save-hook 'highlight-save)
| highlight-save |
helm-swoop–clear-cache |
cat .highlights.org.highlights
((438 454 "Light Salmon" "highlighted") (1014 1031 "Light Salmon" "highlighted"))
Here, we can read the contents and apply the highlights. We set this up on a hook for org-mode, so it will apply them on when we open org-files. You could make this more general if you plan to highlight in code files, for example.
(defun highlight-load ()
"Load and apply highlights."
(interactive)
(setq font-lock-extra-managed-props (delq 'help-echo font-lock-extra-managed-props))
(let ((fname (highlight-save-filename)))
(when (and fname (file-exists-p fname))
(mapcar
(lambda (entry)
(let ((beg (nth 0 entry))
(end (nth 1 entry))
(color (nth 2 entry))
(help-echo (nth 3 entry)))
(set-text-properties
beg end
`(font-lock-face (:background ,color)
help-echo ,help-echo
highlighted t))))
(with-temp-buffer (insert-file-contents fname)
(read (current-buffer)))))))
(add-hook 'org-mode-hook 'highlight-load)
Now, let's outdo ourselves in ridiculosity. We will add a helm-colors selector to give you unprecedented highlighting capability in multicolor magnificence. This function will highlight selected text, or update the color of an existing highlight.
(defun highlight (beg end &optional color)
"Highlight region from BEG to END with COLOR.
COLOR is selected from `helm-colors' when run interactively."
(interactive "r")
(unless (or (get-text-property (point) 'highlighted)
(region-active-p))
(error "No region selected or not on a highlight."))
(unless color
(setq color (s-trim (helm-colors))))
(if (get-text-property (point) 'highlighted)
;; update color
(let ((beg (previous-single-property-change (point) 'highlighted))
(end (next-single-property-change (point) 'highlighted)))
(set-text-properties
beg end
`(font-lock-face (:background ,color)
highlighted t)))
(set-text-properties
beg end
`(font-lock-face (:background ,color)
highlighted t))))
;; For convenience
(global-set-key (kbd "s-h") 'highlight)
highlight
Now, we can conveniently highlight text in whatever color we want. How about list your highlights? After we have highlighted a lot, it might be nice to see a list of these we can click on to find our highlights more quickly.
(defun highlight-list ()
"Make a list of highlighted text in another buffer. "
(interactive)
(let ((cb (current-buffer))
(fname (buffer-file-name))
(hls (mapcar
(lambda (entry)
(list (nth 0 entry)
(buffer-substring (nth 0 entry) (nth 1 entry))))
(highlight-get-highlights))))
(if hls
(progn
(split-window-right)
(switch-to-buffer-other-window "*highlights*") (org-mode)
(setq buffer-read-only nil)
(erase-buffer)
(insert "Click on text to jump to the position.\n\n")
(dolist (s hls)
(let ((map (make-sparse-keymap)))
(define-key map [mouse-1]
`(lambda ()
(interactive)
(find-file ,fname)
(goto-char ,(nth 0 s))))
(insert (propertize
(concat (nth 1 s) "\n")
'local-map map))))
(setq buffer-read-only t))
(message "No highlights found."))))
highlight-list
You probably would like to just select some text with your mouse, and have it highlighted. That requires us to advise the mouse-set-region function.
(defun highlight-green ()
"Highlight region in green."
(interactive)
(highlight (region-beginning) (region-end) "Darkolivegreen1"))
;; create the advice for use later
(defadvice mouse-set-region (after my-highlight () disable)
"Highlight"
(highlight-green))
(defun highlight-mouse-on ()
"Turn on mouse highlighting"
(interactive)
(ad-enable-advice 'mouse-set-region 'after 'my-highlight)
(ad-activate 'mouse-set-region))
(defun highlight-mouse-off ()
(interactive)
(ad-disable-advice 'mouse-set-region 'after 'my-highlight)
(ad-deactivate 'mouse-set-region))
highlight-mouse-off
(defun highlight-picasso-blues ()
(interactive)
(save-excursion
(let ((colors '("PowderBlue"
"Lightskyblue1"
"Lightskyblue2"
"Lightskyblue3"
"Lightskyblue4"))
(beg (region-beginning))
(end (region-end)))
(goto-char beg)
(while (< (point) (- end 1))
(highlight (point) (+ 1 (point))
(nth (mod (- (point) (region-beginning)) (length colors)) colors))
(forward-char)))))
(defun highlight-rainbow ()
(interactive)
(save-excursion
(let ((colors '("Red1"
"Orange1"
"Yellow1"
"Darkolivegreen1"
"Skyblue1"
"Blue1"
"DarkViolet"))
(beg (region-beginning))
(end (region-end)))
(goto-char beg)
(while (< (point) (- end 1))
(highlight (point) (+ 1 (point))
(nth (mod (- (point) (region-beginning)) (length colors)) colors))
(forward-char)))))
=These look cool, but they don't get properly saved. The code that finds the highlights finds the region, but only saves the first color. That means that adjacent highlights of different color will also not be saved correctly.
How about a highlight with your own tooltip? In theory we can set the help-echo property to some text. In practice I have found this tricky because font-lock occasionally erases help-echo properties on re-fontifying. We remove help-echo from a list of properties that are affected by this, but another library may add it back, and there might be some unintended consequences of that. Here we design a function to highlight with a user-defined tooltip.
(defun highlight-note (beg end color &optional note)
"Highlight selected text and add NOTE to it as a tooltip."
(interactive
(list
(region-beginning)
(region-end)
(s-trim (helm-colors))))
(unless note (setq note (read-input "Note: ")))
(unless (region-active-p)
(error "No region selected."))
(set-text-properties
beg end
`(help-echo ,note font-lock-face (:background ,color)
highlighted t)))
(defun highlight-note-edit (new-note)
"Set tooltip of highlight at point to NEW-NOTE."
(interactive (list (read-input "Note: " (get-text-property (point) 'help-echo))))
(let* ((region (button-lock-find-extent (point) 'highlighted))
(beg (car region))
(end (cdr region)))
(put-text-property beg end 'help-echo new-note)))
=highlight-note-edit
==highlight-note-edit
==highlight-note-edit
==highlight-note-edit
=C
Want to get rid of the highlights? We may want to delete one or all. We make a function for each.
(defun highlight-clear ()
"Clear highlight at point."
(interactive)
(when (get-text-property (point) 'highlighted)
(set-text-properties
(next-single-property-change (point) 'highlighted)
(previous-single-property-change (point) 'highlighted)
nil)))
(defun highlight-clear-all ()
"Clear all highlights.
They are really deleted when you save the buffer."
(interactive)
(mapcar
(lambda (entry)
(let ((beg (nth 0 entry))
(end (nth 1 entry)))
(set-text-properties
beg end nil)))
(highlight-get-highlights))
(when (get-buffer "*highlights*")
(kill-buffer "*highlights*")))
highlight-clear-all
Let's define a few convenience functions for common colors, a hydra to quickly select them and bind it to a key for convenience. While we are at it, we add a menu to Org too.
(defun highlight-yellow ()
"Highlight region in yellow."
(interactive)
(highlight (region-beginning) (region-end) "Yellow"))
(defun highlight-blue ()
"Highlight region in blue."
(interactive)
(highlight (region-beginning) (region-end) "LightBlue"))
(defun highlight-pink ()
"Highlight region in pink."
(interactive)
(highlight (region-beginning) (region-end) "Pink"))
(defun highlight-green ()
"Highlight region in green."
(interactive)
(highlight (region-beginning) (region-end) "Darkolivegreen1"))
(defhydra highlighter (:color blue) "highlighter"
("b" highlight-blue "blue")
("g" highlight-green "Green")
("p" highlight-pink "Pink")
;; define as many special colors as you like.
("s" (highlight (region-beginning) (region-end) "Lightsalmon1") "Salmon")
("y" highlight-yellow "yellow")
("c" highlight "Choose color")
("n" (highlight-note (region-beginning) (region-end) "Thistle") "Note")
("N" highlight-note "Note (c)")
("m" highlight-mouse-on "Mouse")
("M" highlight-mouse-off "Mouse off")
("e" highlight-note-edit "Edit note")
("l" highlight-list "List highlights")
("r" highlight-load "Reload")
("S" highlight-save "Save")
("d" highlight-clear "Delete")
("D" highlight-clear-all "Delete All"))
(easy-menu-change
'("Org") "highlighter"
'(["Highlight" highlight]
["Highlight (B)" highlight-blue]
["Highlight (G)" highlight-green]
["Highlight (P)" highlight-pink]
["Highlight (Y)" highlight-yellow]
["Highlight note" highlight-note]
["List highlights" highlight-list]
["Delete highlight" highlight-clear]
["Delete highlights" highlight-clear-all])
"Show/Hide")
(global-set-key (kbd "s-h") 'highlighter/body)
highlighter/body
1 Known limitations
The tooltips seem especially fragile, and if there is code that undoes the removal of help-echo from font-lock-extra-managed-props, it seems possible they would easily get lost. I wouldn't use them a lot without a lot of testing. You have to rely on the hook functions defined to keep the highlights synchronized between the buffer and the external highlight file. If you were to rename a file externally, e.g. in the OS, or with a shell command, then the highlights will be lost unless you also rename the external file.
Highlights are not robust enough to survive refiling an org-mode section from one file to another. Personally I don't see these as too big a problem; I don't put a lot of value of highlights, but I can see it being pretty annoying to lose them!
Still, if you want to give this a try, you can use the code here: highlights.el . You should bind the functions to whatever keys you want. Also, it is setup to only work for org-mode. I am not sure what the best hook to use for any file might be. Maybe find-file-hook.
Copyright (C) 2015 by John Kitchin. See the License for information about copying.
org-mode source
Org-mode version = 8.2.10
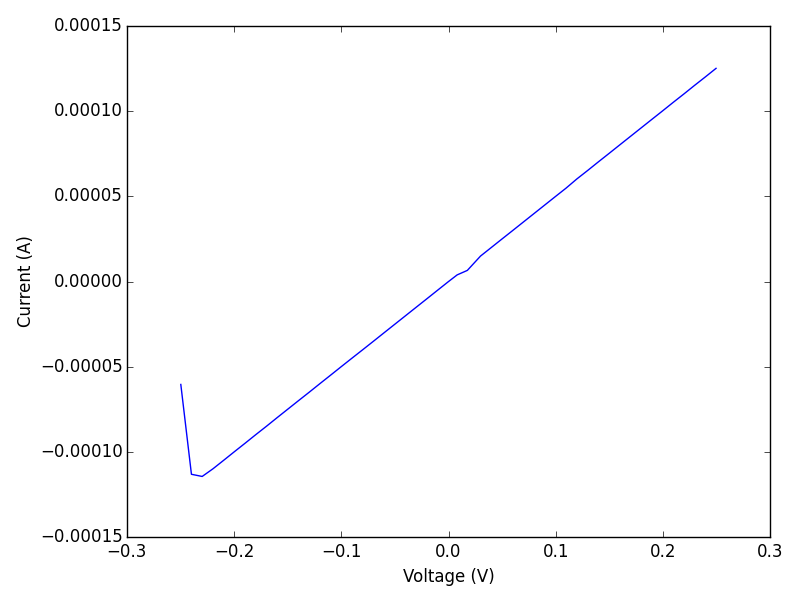 Well, there you have it. Possibly the first Gamry Ref600 to ever have been driven from a Mac ;) Let me be more explicit about that; I could also run this from Linux, an iPad, etc… You could do this in a browser, or in an IPython notebook, or in Matlab, among many other possibilities. You could write a script in perl, shell, ruby, emacs-lisp, or any other language that supports http requests.
Well, there you have it. Possibly the first Gamry Ref600 to ever have been driven from a Mac ;) Let me be more explicit about that; I could also run this from Linux, an iPad, etc… You could do this in a browser, or in an IPython notebook, or in Matlab, among many other possibilities. You could write a script in perl, shell, ruby, emacs-lisp, or any other language that supports http requests.