Matlab post
Usually, the numerical ODE solvers in python work well with the standard settings. Sometimes they do not, and it is not always obvious they have not worked! Part of using a tool like python is checking how well your solution really worked. We use an example of integrating an ODE that defines the van der Waal equation of an ideal gas here.
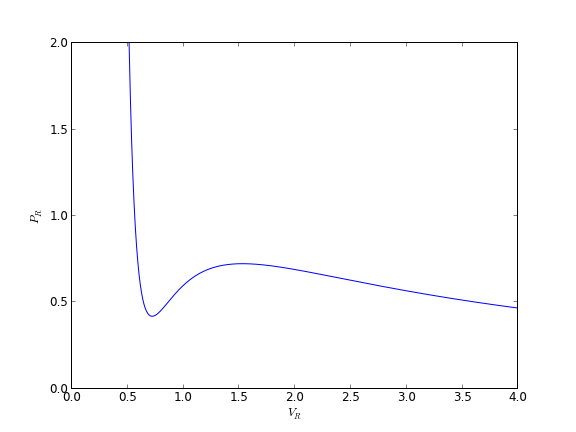
we plot the analytical solution to the van der waal equation in reduced form here.
import numpy as np
import matplotlib.pyplot as plt
Tr = 0.9
Vr = np.linspace(0.34,4,1000)
#analytical equation for Pr
Prfh = lambda Vr: 8.0 / 3.0 * Tr / (Vr - 1.0 / 3.0) - 3.0 / (Vr**2)
Pr = Prfh(Vr) # evaluated on our reduced volume vector.
# Plot the EOS
plt.plot(Vr,Pr)
plt.ylim([0, 2])
plt.xlabel('$V_R$')
plt.ylabel('$P_R$')
plt.savefig('images/ode-vw-1.png')
plt.show()
>>> >>> >>> >>> >>> ... >>> >>> >>> ... [<matplotlib.lines.Line2D object at 0x1c5a3550>]
(0, 2)
<matplotlib.text.Text object at 0x1c22f750>
<matplotlib.text.Text object at 0x1d4e0750>
we want an equation for dPdV, which we will integrate we use symbolic math to do the derivative for us.
from sympy import diff, Symbol
Vrs = Symbol('Vrs')
Prs = 8.0 / 3.0 * Tr / (Vrs - 1.0/3.0) - 3.0/(Vrs**2)
print diff(Prs,Vrs)
>>> -2.4/(Vrs - 0.333333333333333)**2 + 6.0/Vrs**3
Now, we solve the ODE. We will specify a large relative tolerance criteria (Note the default is much smaller than what we show here).
from scipy.integrate import odeint
def myode(Pr, Vr):
dPrdVr = -2.4/(Vr - 0.333333333333333)**2 + 6.0/Vr**3
return dPrdVr
Vspan = np.linspace(0.334, 4)
Po = Prfh(Vspan[0])
P = odeint(myode, Po, Vspan, rtol=1e-4)
# Plot the EOS
plt.plot(Vr,Pr) # analytical solution
plt.plot(Vspan, P[:,0], 'r.')
plt.ylim([0, 2])
plt.xlabel('$V_R$')
plt.ylabel('$P_R$')
plt.savefig('images/ode-vw-2.png')
plt.show()
... >>> >>> >>> >>> ... [<matplotlib.lines.Line2D object at 0x1d4f3b90>]
[<matplotlib.lines.Line2D object at 0x2ac47518e710>]
(0, 2)
<matplotlib.text.Text object at 0x1c238fd0>
<matplotlib.text.Text object at 0x1c22af10>
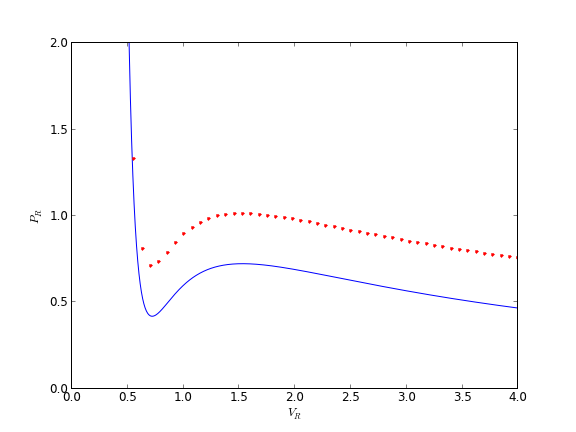
You can see there is disagreement between the analytical solution and numerical solution. The origin of this problem is accuracy at the initial condition, where the derivative is extremely large.
-53847.3437818
We can increase the tolerance criteria to get a better answer. The defaults in odeint are actually set to 1.49012e-8.
Vspan = np.linspace(0.334, 4)
Po = Prfh(Vspan[0])
P = odeint(myode, Po, Vspan)
# Plot the EOS
plt.plot(Vr,Pr) # analytical solution
plt.plot(Vspan, P[:,0], 'r.')
plt.ylim([0, 2])
plt.xlabel('$V_R$')
plt.ylabel('$P_R$')
plt.savefig('images/ode-vw-3.png')
plt.show()
>>> ... [<matplotlib.lines.Line2D object at 0x1d4dbf10>]
[<matplotlib.lines.Line2D object at 0x1c6e5550>]
(0, 2)
<matplotlib.text.Text object at 0x1d4e31d0>
<matplotlib.text.Text object at 0x1d9d3710>
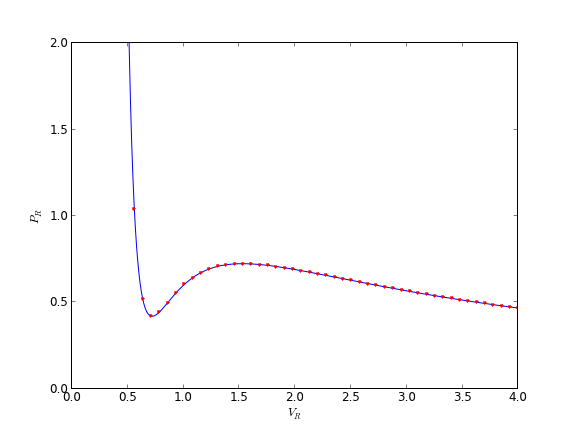
The problem here was the derivative value varied by four orders of magnitude over the integration range, so the default tolerances were insufficient to accurately estimate the numerical derivatives over that range. Tightening the tolerances helped resolve that problem. Another approach might be to split the integration up into different regions. For instance, if instead of starting at Vr = 0.34, which is very close to a sigularity in the van der waal equation at Vr = 1/3, if you start at Vr = 0.5, the solution integrates just fine with the standard tolerances.
Copyright (C) 2013 by John Kitchin. See the License for information about copying.
org-mode source