Plug flow reactor with a pressure drop
Posted February 18, 2013 at 09:00 AM | categories: ode | tags: reaction engineering, fluids
Updated March 06, 2013 at 04:39 PM
If there is a pressure drop in a plug flow reactor, 1 there are two equations needed to determine the exit conversion: one for the conversion, and one from the pressure drop.
\begin{eqnarray} \frac{dX}{dW} &=& \frac{k'}{F_A0} \left ( \frac{1-X}{1 + \epsilon X} \right) y\\ \frac{dX}{dy} &=& -\frac{\alpha (1 + \epsilon X)}{2y} \end{eqnarray}Here is how to integrate these equations numerically in python.
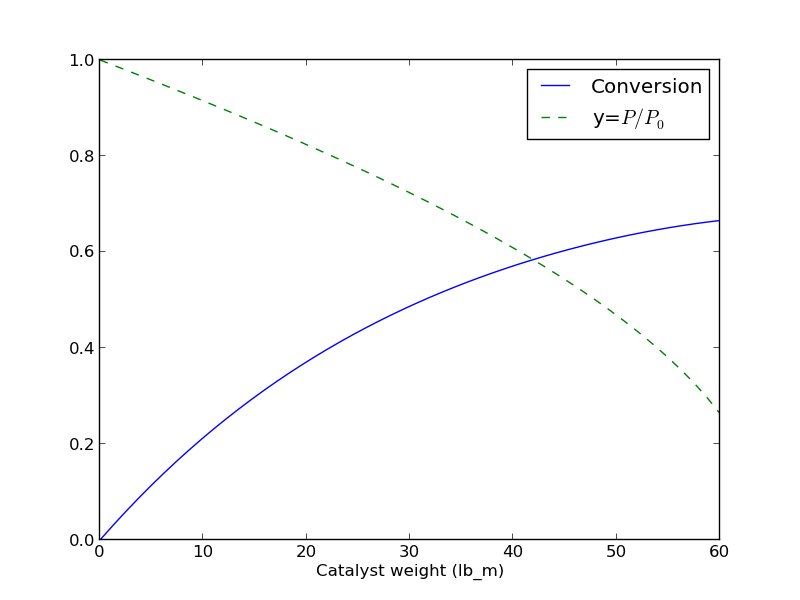
import numpy as np from scipy.integrate import odeint import matplotlib.pyplot as plt kprime = 0.0266 Fa0 = 1.08 alpha = 0.0166 epsilon = -0.15 def dFdW(F, W): 'set of ODEs to integrate' X = F[0] y = F[1] dXdW = kprime / Fa0 * (1-X) / (1 + epsilon*X) * y dydW = -alpha * (1 + epsilon * X) / (2 * y) return [dXdW, dydW] Wspan = np.linspace(0,60) X0 = 0.0 y0 = 1.0 F0 = [X0, y0] sol = odeint(dFdW, F0, Wspan) # now plot the results plt.plot(Wspan, sol[:,0], label='Conversion') plt.plot(Wspan, sol[:,1], 'g--', label='y=$P/P_0$') plt.legend(loc='best') plt.xlabel('Catalyst weight (lb_m)') plt.savefig('images/2013-01-08-pdrop.png')
Here is the resulting figure.
Copyright (C) 2013 by John Kitchin. See the License for information about copying.