2022 in a nutshell
Posted January 01, 2023 at 01:52 PM | categories: news | tags:
Table of Contents
2022 was an interesting one. I still did all of my teaching remotely, but spent more time going in to the office for meetings, and have gotten back to professional travel for meetings.
1. Research group accomplishments
Minjie Liu and Yilin Yang both defended their PhDs and graduated in 2022. Luyang Liu, Ananya Srivastava and Karan Waghela all completed their MS degrees and graduated in 2022 also. Congratulations to all of them!
Maya Bhat and I participated in an iCorps workshop on a concept around design of experiments.
We welcomed seven PhD students from the Ulissi group into the group while he is on leave at Meta. We also welcomed two new first year PhD students who will begin new collaborative projects with Carl Laird and Andy Gellman. The group is suddenly quite large!
2. Publications
Our work this past year was divided in a few efforts. We had some work in method development, e.g. in uncertainty quantification, automatic differentiation, and vector search.
- Yang, Y., Achar, S. K., & Kitchin, J. R. (2022). Evaluation of the degree of rate control via automatic differentiation. AIChE Journal, 68(6). http://dx.doi.org/10.1002/aic.17653
- Yang, Y., Liu, M., & Kitchin, J. R. (2022). Neural network embeddings based similarity search method for atomistic systems. Digital Discovery. http://dx.doi.org/10.1039/d2dd00055e
- Zhan, N., & Kitchin, J. R. (2022). Model-specific to model-general uncertainty for physical properties. Industrial & Engineering Chemistry Research, 1–04706. http://dx.doi.org/10.1021/acs.iecr.1c04706
This collaborative work on large catalyst models is was especially exciting. Stay tuned for many advances in this area in 2023.
- Kolluru, A., Shuaibi, M., Palizhati, A., Shoghi, N., Das, A., Wood, B., Zitnick, C. L., … (2022). Open challenges in developing generalizable large-scale machine-learning models for catalyst discovery. ACS Catalysis, 12(14), 8572–8581. http://dx.doi.org/10.1021/acscatal.2c02291
We also wrote some collaborative papers on our work in high-throughput discovery of hydrogen evolution catalysts and segregation.
- Broderick, K., Lopato, E., Wander, B., Bernhard, S., Kitchin, J., & Ulissi, Z. (2022). Identifying limitations in screening high-throughput photocatalytic bimetallic nanoparticles with machine-learned hydrogen adsorptions. Applied Catalysis B: Environmental, 121959. http://dx.doi.org/10.1016/j.apcatb.2022.121959
- Bhat, M., Lopato, E., Simon, Z. C., Millstone, J. E., Bernhard, S., & Kitchin, J. R. (2022). Accelerated optimization of pure metal and ligand compositions for light-driven hydrogen production. Reaction Chemistry & Engineering. http://dx.doi.org/10.1039/d1re00441g
- Yilin Yang, Zhitao Guo, Andrew Gellman and John Kitchin, Simulating segregation in a ternary Cu-Pd-Au alloy with density functional theory, machine learning and Monte Carlo simulations, J. Phys. Chem. C, 126, 4, 1800-1808. (2022). https://pubs.acs.org/doi/abs/10.1021/acs.jpcc.1c09647
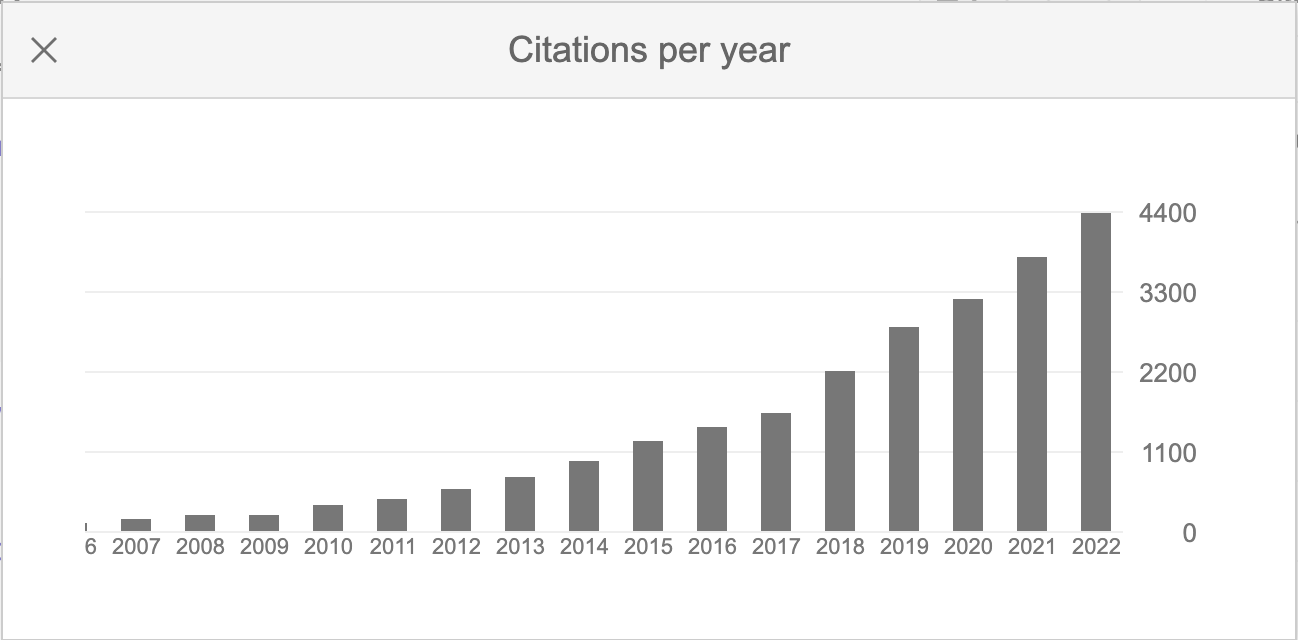
It was another big year in citations for us (https://scholar.google.com/citations?user=jD_4h7sAAAAJ)!
3. Point Breeze Publishing, LLC
I started a publishing company this year Point Breeze Publishing, LLC. It is a way for me to sell booklets on using Python in Science and Engineering and to sustain the effort it takes to produce these. It has been a modest success so far, with about a dozen booklets that can help anyone get started from basic Python usage through data science and machine learning and design of experiments. For reading this, you can get 50% off all purchases with checkout code 2022-nutshell. Check it out, and leave a review if you get anything!
I am still working out what the next steps for this are. I have written most of the pycse content I had in mind now, 400+ pages of it. I would like to get these booklets in the hands of more students, and my stealthy advertising scheme on Twitter and YouTube has not made that happen yet. I have some ideas around molecular simulation, maybe a reboot of the DFT-book?, maybe something around scimax? Who knows, stay tuned!
4. Outlook for 2023
There will be lots of changes for the Kitchin Research Group in 2023. We had a massive growth at the end of 2022 as we welcomed many members of the Ulissi research group into our group while he is on leave at Meta for 2023. Last summer we had one PhD student and three MS students. Now we have 10 PhD students. That means we will start a lot of new research directions in large catalyst models and everything they enable. We have started several collaborations in the area of design of experiments, and look forward to seeing these grow. It should be exciting!
Copyright (C) 2023 by John Kitchin. See the License for information about copying.
Org-mode version = 9.5.5