Using org-entries like a database
Posted August 21, 2014 at 08:45 PM | categories: orgmode, dft | tags:
Updated August 22, 2014 at 10:27 AM
Table of Contents
- 1. Generating the entries
- 1.1. 10/eos-exp/f+0 10 eos_dash_exp f_plus_0
- 1.2. 10/eos-exp/f+10 10 eos_dash_exp f_plus_10
- 1.3. 10/eos-exp/f+12 10 eos_dash_exp f_plus_12
- 1.4. 10/eos-exp/f+15 10 eos_dash_exp f_plus_15
- 1.5. 10/eos-exp/f+2 10 eos_dash_exp f_plus_2
- 1.6. 10/eos-exp/f+4 10 eos_dash_exp f_plus_4
- 1.7. 10/eos-exp/f+6 10 eos_dash_exp f_plus_6
- 1.8. 10/eos-exp/f+8 10 eos_dash_exp f_plus_8
- 1.9. 10/eos-exp/f-10 10 eos_dash_exp f_dash_10
- 1.10. 10/eos-exp/f-12 10 eos_dash_exp f_dash_12
- 1.11. 10/eos-exp/f-15 10 eos_dash_exp f_dash_15
- 1.12. 10/eos-exp/f-2 10 eos_dash_exp f_dash_2
- 1.13. 10/eos-exp/f-4 10 eos_dash_exp f_dash_4
- 1.14. 10/eos-exp/f-6 10 eos_dash_exp f_dash_6
- 1.15. 10/eos-exp/f-8 10 eos_dash_exp f_dash_8
- 1.16. 11/eos-exp/f+0 11 eos_dash_exp f_plus_0
- 1.17. 11/eos-exp/f+10 11 eos_dash_exp f_plus_10
- 1.18. 11/eos-exp/f+12 11 eos_dash_exp f_plus_12
- 1.19. 11/eos-exp/f+15 11 eos_dash_exp f_plus_15
- 1.20. 11/eos-exp/f+2 11 eos_dash_exp f_plus_2
- 1.21. 11/eos-exp/f+4 11 eos_dash_exp f_plus_4
- 1.22. 11/eos-exp/f+6 11 eos_dash_exp f_plus_6
- 1.23. 11/eos-exp/f+8 11 eos_dash_exp f_plus_8
- 1.24. 11/eos-exp/f-10 11 eos_dash_exp f_dash_10
- 1.25. 11/eos-exp/f-12 11 eos_dash_exp f_dash_12
- 1.26. 11/eos-exp/f-15 11 eos_dash_exp f_dash_15
- 1.27. 11/eos-exp/f-2 11 eos_dash_exp f_dash_2
- 1.28. 11/eos-exp/f-4 11 eos_dash_exp f_dash_4
- 1.29. 11/eos-exp/f-6 11 eos_dash_exp f_dash_6
- 1.30. 11/eos-exp/f-8 11 eos_dash_exp f_dash_8
- 2. Tabulating the results in column mode
- 3. org-map-entries to get the data
- 4. Getting specific data from a calculation
- 5. Summary
I recently added an org export to jasp which provides an org-mode representation of a Vasp calculation. It is still a work in progress, but the main ideas are developed. The idea is to represent a calculation as an org-mode headline with tabular data, properties and tags. This will make the representation human readable, but machine accessible. Here I examine how we could use it.
1 Generating the entries
We will loop over a set of calculations that define the equation of state for two different Al-Ni alloy structures. For each calculation this code generates a level 3 headline with some tags derived from the calculation directory. I chose level 3 so they would all be nested in this level 2 headline. We write out two sets of data because later we will selectively get portions of the data.
from jasp import * from jasp.utils import get_jasp_dirs for d in get_jasp_dirs('10'): with jasp(d) as calc: print calc.org(level=3) for d in get_jasp_dirs('11'): with jasp(d) as calc: print calc.org(level=3)
1.1 10/eos-exp/f+0 10 eos_dash_exp f_plus_0
| x | y | z | |
|---|---|---|---|
| A0 | -1.98408 | 0.0 | -1.98408 |
| A1 | 0.0 | -1.98408 | 1.98408 |
| A2 | -3.53625 | 3.53625 | 3.53625 |
| species | x | y | z | fx | fy | fz | free x | free y | free z |
|---|---|---|---|---|---|---|---|---|---|
| Al | -3.77549 | 1.79141 | 1.79141 | 0.000773 | -0.000773 | -0.000773 | T | T | T |
| Al | -1.74484 | -0.23924 | 1.74484 | -0.000773 | 0.000773 | 0.000773 | T | T | T |
| Ni | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | T | T | T |
| Parameter | Value |
|---|---|
| nbands | 25 |
| encut | 375.0 |
| sigma | 0.1 |
| prec | Normal |
| kpts | [12, 12, 5] |
| xc | PBE |
| txt | - |
| gamma | True |
| Ni | potpawPBE/Ni/POTCAR | 6c814bc7144b98435771aded093fb4ebe527acd9 |
| Al | potpawPBE/Al/POTCAR | c8d9ecb0b6ebec0256c5f5072cee4de6a046dac2 |
1.2 10/eos-exp/f+10 10 eos_dash_exp f_plus_10
| x | y | z | |
|---|---|---|---|
| A0 | -2.04813 | 0.0 | -2.04813 |
| A1 | 0.0 | -2.04813 | 2.04813 |
| A2 | -3.6504 | 3.6504 | 3.6504 |
| species | x | y | z | fx | fy | fz | free x | free y | free z |
|---|---|---|---|---|---|---|---|---|---|
| Al | -3.85098179507 | 1.84576178527 | 1.83645153581 | -0.012598 | 0.014577 | -0.007395 | T | T | T |
| Al | -1.7283215185 | -0.279882546245 | 1.73598892646 | 0.000817 | 0.006771 | 0.025221 | T | T | T |
| Ni | -5.63955668642 | 1.58856076097 | 3.61420953773 | 0.011781 | -0.021349 | -0.017826 | T | T | T |
| Parameter | Value |
|---|---|
| nbands | 25 |
| encut | 375.0 |
| sigma | 0.1 |
| prec | Normal |
| kpts | [12, 12, 5] |
| xc | PBE |
| txt | - |
| gamma | True |
| Ni | potpawPBE/Ni/POTCAR | 6c814bc7144b98435771aded093fb4ebe527acd9 |
| Al | potpawPBE/Al/POTCAR | c8d9ecb0b6ebec0256c5f5072cee4de6a046dac2 |
1.3 10/eos-exp/f+12 10 eos_dash_exp f_plus_12
| x | y | z | |
|---|---|---|---|
| A0 | -2.06046 | 0.0 | -2.06046 |
| A1 | 0.0 | -2.06046 | 2.06046 |
| A2 | -3.67239 | 3.67239 | 3.67239 |
| species | x | y | z | fx | fy | fz | free x | free y | free z |
|---|---|---|---|---|---|---|---|---|---|
| Al | -3.86530078437 | 1.85683453967 | 1.84560971979 | -0.017615 | 0.018377 | -0.00588 | T | T | T |
| Al | -1.72555029376 | -0.287469117251 | 1.73429429921 | 0.009139 | -0.000441 | 0.020494 | T | T | T |
| Ni | -5.66232892187 | 1.59473457758 | 3.628735981 | 0.008476 | -0.017936 | -0.014613 | T | T | T |
| Parameter | Value |
|---|---|
| nbands | 25 |
| encut | 375.0 |
| sigma | 0.1 |
| prec | Normal |
| kpts | [12, 12, 5] |
| xc | PBE |
| txt | - |
| gamma | True |
| Ni | potpawPBE/Ni/POTCAR | 6c814bc7144b98435771aded093fb4ebe527acd9 |
| Al | potpawPBE/Al/POTCAR | c8d9ecb0b6ebec0256c5f5072cee4de6a046dac2 |
1.4 10/eos-exp/f+15 10 eos_dash_exp f_plus_15
| x | y | z | |
|---|---|---|---|
| A0 | -2.0787 | 0.0 | -2.0787 |
| A1 | 0.0 | -2.0787 | 2.0787 |
| A2 | -3.70489 | 3.70489 | 3.70489 |
| species | x | y | z | fx | fy | fz | free x | free y | free z |
|---|---|---|---|---|---|---|---|---|---|
| Al | -3.88717652995 | 1.87379242704 | 1.86009271779 | -0.022309 | 0.021755 | -0.006413 | T | T | T |
| Al | -1.72325929808 | -0.297739728075 | 1.73317610987 | 0.033819 | -0.022553 | -0.000329 | T | T | T |
| Ni | -5.69348417197 | 1.60230730103 | 3.64787117234 | -0.01151 | 0.000799 | 0.006742 | T | T | T |
| Parameter | Value |
|---|---|
| nbands | 25 |
| encut | 375.0 |
| sigma | 0.1 |
| prec | Normal |
| kpts | [12, 12, 5] |
| xc | PBE |
| txt | - |
| gamma | True |
| Ni | potpawPBE/Ni/POTCAR | 6c814bc7144b98435771aded093fb4ebe527acd9 |
| Al | potpawPBE/Al/POTCAR | c8d9ecb0b6ebec0256c5f5072cee4de6a046dac2 |
1.5 10/eos-exp/f+2 10 eos_dash_exp f_plus_2
| x | y | z | |
|---|---|---|---|
| A0 | -1.99722 | 0.0 | -1.99722 |
| A1 | 0.0 | -1.99722 | 1.99722 |
| A2 | -3.55967 | 3.55967 | 3.55967 |
| species | x | y | z | fx | fy | fz | free x | free y | free z |
|---|---|---|---|---|---|---|---|---|---|
| Al | -3.78897262915 | 1.80298810692 | 1.80238679997 | -0.00052 | -0.01106 | -0.017256 | T | T | T |
| Al | -1.74283113391 | -0.243416998758 | 1.743431885 | 0.011316 | -0.021883 | -0.005492 | T | T | T |
| Ni | -5.54541623693 | 1.55504889184 | 3.55010131503 | -0.010796 | 0.032943 | 0.022747 | T | T | T |
| Parameter | Value |
|---|---|
| nbands | 25 |
| encut | 375.0 |
| sigma | 0.1 |
| prec | Normal |
| kpts | [12, 12, 5] |
| xc | PBE |
| txt | - |
| gamma | True |
| Ni | potpawPBE/Ni/POTCAR | 6c814bc7144b98435771aded093fb4ebe527acd9 |
| Al | potpawPBE/Al/POTCAR | c8d9ecb0b6ebec0256c5f5072cee4de6a046dac2 |
1.6 10/eos-exp/f+4 10 eos_dash_exp f_plus_4
| x | y | z | |
|---|---|---|---|
| A0 | -2.01019 | 0.0 | -2.01019 |
| A1 | 0.0 | -2.01019 | 2.01019 |
| A2 | -3.58278 | 3.58278 | 3.58278 |
| species | x | y | z | fx | fy | fz | free x | free y | free z |
|---|---|---|---|---|---|---|---|---|---|
| Al | -3.80137127251 | 1.81395280614 | 1.81276515721 | -0.021611 | -0.002318 | -0.01379 | T | T | T |
| Al | -1.73979870776 | -0.248609560277 | 1.74098667865 | 0.017491 | -0.038446 | -0.007248 | T | T | T |
| Ni | -5.57213001973 | 1.55941675414 | 3.56527816415 | 0.004121 | 0.040764 | 0.021038 | T | T | T |
| Parameter | Value |
|---|---|
| nbands | 25 |
| encut | 375.0 |
| sigma | 0.1 |
| prec | Normal |
| kpts | [12, 12, 5] |
| xc | PBE |
| txt | - |
| gamma | True |
| Ni | potpawPBE/Ni/POTCAR | 6c814bc7144b98435771aded093fb4ebe527acd9 |
| Al | potpawPBE/Al/POTCAR | c8d9ecb0b6ebec0256c5f5072cee4de6a046dac2 |
1.7 10/eos-exp/f+6 10 eos_dash_exp f_plus_6
| x | y | z | |
|---|---|---|---|
| A0 | -2.02299 | 0.0 | -2.02299 |
| A1 | 0.0 | -2.02299 | 2.02299 |
| A2 | -3.60561 | 3.60561 | 3.60561 |
| species | x | y | z | fx | fy | fz | free x | free y | free z |
|---|---|---|---|---|---|---|---|---|---|
| Al | -3.81674368134 | 1.82268911017 | 1.81846628092 | -0.03365 | 0.021418 | 0.006392 | T | T | T |
| Al | -1.73592646487 | -0.259496276667 | 1.73977985143 | 0.007575 | -0.01713 | 0.007756 | T | T | T |
| Ni | -5.5962598538 | 1.5715971665 | 3.58361386765 | 0.026075 | -0.004288 | -0.014148 | T | T | T |
| Parameter | Value |
|---|---|
| nbands | 25 |
| encut | 375.0 |
| sigma | 0.1 |
| prec | Normal |
| kpts | [12, 12, 5] |
| xc | PBE |
| txt | - |
| gamma | True |
| Ni | potpawPBE/Ni/POTCAR | 6c814bc7144b98435771aded093fb4ebe527acd9 |
| Al | potpawPBE/Al/POTCAR | c8d9ecb0b6ebec0256c5f5072cee4de6a046dac2 |
1.8 10/eos-exp/f+8 10 eos_dash_exp f_plus_8
| x | y | z | |
|---|---|---|---|
| A0 | -2.03564 | 0.0 | -2.03564 |
| A1 | 0.0 | -2.03564 | 2.03564 |
| A2 | -3.62814 | 3.62814 | 3.62814 |
| species | x | y | z | fx | fy | fz | free x | free y | free z |
|---|---|---|---|---|---|---|---|---|---|
| Al | -3.83039067392 | 1.83353256518 | 1.82788023062 | -0.042201 | 0.025424 | 0.00725 | T | T | T |
| Al | -1.73261729052 | -0.266893248067 | 1.73758502884 | 0.008197 | -0.020125 | 0.010395 | T | T | T |
| Ni | -5.62110203556 | 1.57803068288 | 3.59892474053 | 0.034004 | -0.005299 | -0.017645 | T | T | T |
| Parameter | Value |
|---|---|
| nbands | 25 |
| encut | 375.0 |
| sigma | 0.1 |
| prec | Normal |
| kpts | [12, 12, 5] |
| xc | PBE |
| txt | - |
| gamma | True |
| Ni | potpawPBE/Ni/POTCAR | 6c814bc7144b98435771aded093fb4ebe527acd9 |
| Al | potpawPBE/Al/POTCAR | c8d9ecb0b6ebec0256c5f5072cee4de6a046dac2 |
1.9 10/eos-exp/f-10 10 eos_dash_exp f_dash_10
| x | y | z | |
|---|---|---|---|
| A0 | -1.91561 | 0.0 | -1.91561 |
| A1 | 0.0 | -1.91561 | 1.91561 |
| A2 | -3.41421 | 3.41421 | 3.41421 |
| species | x | y | z | fx | fy | fz | free x | free y | free z |
|---|---|---|---|---|---|---|---|---|---|
| Al | -3.69513184604 | 1.73676514888 | 1.75200457401 | -0.00015 | -0.016008 | 0.015593 | T | T | T |
| Al | -1.76033286748 | -0.198966401698 | 1.74405584289 | 0.003022 | -0.015555 | -0.029074 | T | T | T |
| Ni | -0.0648652864789 | 0.0143712528207 | 0.040189583095 | -0.002872 | 0.031562 | 0.013481 | T | T | T |
| Parameter | Value |
|---|---|
| nbands | 25 |
| encut | 375.0 |
| sigma | 0.1 |
| prec | Normal |
| kpts | [12, 12, 5] |
| xc | PBE |
| txt | - |
| gamma | True |
| Ni | potpawPBE/Ni/POTCAR | 6c814bc7144b98435771aded093fb4ebe527acd9 |
| Al | potpawPBE/Al/POTCAR | c8d9ecb0b6ebec0256c5f5072cee4de6a046dac2 |
1.10 10/eos-exp/f-12 10 eos_dash_exp f_dash_12
| x | y | z | |
|---|---|---|---|
| A0 | -1.90131 | 0.0 | -1.90131 |
| A1 | 0.0 | -1.90131 | 1.90131 |
| A2 | -3.38873 | 3.38873 | 3.38873 |
| species | x | y | z | fx | fy | fz | free x | free y | free z |
|---|---|---|---|---|---|---|---|---|---|
| Al | -3.67627852264 | 1.72365573425 | 1.74704265204 | -0.021096 | -0.003148 | 0.019104 | T | T | T |
| Al | -1.76353019483 | -0.189393782717 | 1.73913926696 | -0.006667 | -0.016634 | -0.009534 | T | T | T |
| Ni | -0.0805212825263 | 0.0179080484715 | 0.050068081002 | 0.027763 | 0.019782 | -0.00957 | T | T | T |
| Parameter | Value |
|---|---|
| nbands | 25 |
| encut | 375.0 |
| sigma | 0.1 |
| prec | Normal |
| kpts | [12, 12, 5] |
| xc | PBE |
| txt | - |
| gamma | True |
| Ni | potpawPBE/Ni/POTCAR | 6c814bc7144b98435771aded093fb4ebe527acd9 |
| Al | potpawPBE/Al/POTCAR | c8d9ecb0b6ebec0256c5f5072cee4de6a046dac2 |
1.11 10/eos-exp/f-15 10 eos_dash_exp f_dash_15
| x | y | z | |
|---|---|---|---|
| A0 | -1.87946 | 0.0 | -1.87946 |
| A1 | 0.0 | -1.87946 | 1.87946 |
| A2 | -3.34978 | 3.34978 | 3.34978 |
| species | x | y | z | fx | fy | fz | free x | free y | free z |
|---|---|---|---|---|---|---|---|---|---|
| Al | -3.65183912264 | 1.70528638812 | 1.73537994296 | -0.037051 | 0.019573 | 0.044452 | T | T | T |
| Al | -1.76884279383 | -0.178373914577 | 1.73770842538 | 0.010475 | -0.025221 | -0.029082 | T | T | T |
| Ni | -0.0996480835307 | 0.0252575264549 | 0.0631616316537 | 0.026577 | 0.005648 | -0.01537 | T | T | T |
| Parameter | Value |
|---|---|
| nbands | 25 |
| encut | 375.0 |
| sigma | 0.1 |
| prec | Normal |
| kpts | [12, 12, 5] |
| xc | PBE |
| txt | - |
| gamma | True |
| Ni | potpawPBE/Ni/POTCAR | 6c814bc7144b98435771aded093fb4ebe527acd9 |
| Al | potpawPBE/Al/POTCAR | c8d9ecb0b6ebec0256c5f5072cee4de6a046dac2 |
1.12 10/eos-exp/f-2 10 eos_dash_exp f_dash_2
| x | y | z | |
|---|---|---|---|
| A0 | -1.97076 | 0.0 | -1.97076 |
| A1 | 0.0 | -1.97076 | 1.97076 |
| A2 | -3.51252 | 3.51252 | 3.51252 |
| species | x | y | z | fx | fy | fz | free x | free y | free z |
|---|---|---|---|---|---|---|---|---|---|
| Al | -3.76146938751 | 1.7792636125 | 1.77972964831 | 0.002765 | 0.010416 | 0.018609 | T | T | T |
| Al | -1.74832410416 | -0.234109027383 | 1.74785846541 | -0.002339 | 0.016333 | -0.006172 | T | T | T |
| Ni | -0.0105365083339 | 0.00701541488599 | 0.00866188628634 | -0.000426 | -0.026749 | -0.012437 | T | T | T |
| Parameter | Value |
|---|---|
| nbands | 25 |
| encut | 375.0 |
| sigma | 0.1 |
| prec | Normal |
| kpts | [12, 12, 5] |
| xc | PBE |
| txt | - |
| gamma | True |
| Ni | potpawPBE/Ni/POTCAR | 6c814bc7144b98435771aded093fb4ebe527acd9 |
| Al | potpawPBE/Al/POTCAR | c8d9ecb0b6ebec0256c5f5072cee4de6a046dac2 |
1.13 10/eos-exp/f-4 10 eos_dash_exp f_dash_4
| x | y | z | |
|---|---|---|---|
| A0 | -1.95726 | 0.0 | -1.95726 |
| A1 | 0.0 | -1.95726 | 1.95726 |
| A2 | -3.48846 | 3.48846 | 3.48846 |
| species | x | y | z | fx | fy | fz | free x | free y | free z |
|---|---|---|---|---|---|---|---|---|---|
| Al | -3.74916407088 | 1.76890327148 | 1.76974035279 | 0.025717 | 0.002077 | 0.019316 | T | T | T |
| Al | -1.75040151275 | -0.230732012577 | 1.74956509347 | -0.018564 | 0.049532 | 8.6e-05 | T | T | T |
| Ni | -0.0207644163691 | 0.0139987410966 | 0.0169445537375 | -0.007153 | -0.051609 | -0.019402 | T | T | T |
| Parameter | Value |
|---|---|
| nbands | 25 |
| encut | 375.0 |
| sigma | 0.1 |
| prec | Normal |
| kpts | [12, 12, 5] |
| xc | PBE |
| txt | - |
| gamma | True |
| Ni | potpawPBE/Ni/POTCAR | 6c814bc7144b98435771aded093fb4ebe527acd9 |
| Al | potpawPBE/Al/POTCAR | c8d9ecb0b6ebec0256c5f5072cee4de6a046dac2 |
1.14 10/eos-exp/f-6 10 eos_dash_exp f_dash_6
| x | y | z | |
|---|---|---|---|
| A0 | -1.94358 | 0.0 | -1.94358 |
| A1 | 0.0 | -1.94358 | 1.94358 |
| A2 | -3.46406 | 3.46406 | 3.46406 |
| species | x | y | z | fx | fy | fz | free x | free y | free z |
|---|---|---|---|---|---|---|---|---|---|
| Al | -3.73031485721 | 1.75693465653 | 1.76095910776 | -0.000377 | 0.017266 | 0.043872 | T | T | T |
| Al | -1.75673108966 | -0.217813140979 | 1.75241474358 | 0.034718 | -0.014395 | -0.060867 | T | T | T |
| Ni | -0.0332840531202 | 0.0130484844485 | 0.0228761486573 | -0.034341 | -0.002871 | 0.016995 | T | T | T |
| Parameter | Value |
|---|---|
| nbands | 25 |
| encut | 375.0 |
| sigma | 0.1 |
| prec | Normal |
| kpts | [12, 12, 5] |
| xc | PBE |
| txt | - |
| gamma | True |
| Ni | potpawPBE/Ni/POTCAR | 6c814bc7144b98435771aded093fb4ebe527acd9 |
| Al | potpawPBE/Al/POTCAR | c8d9ecb0b6ebec0256c5f5072cee4de6a046dac2 |
1.15 10/eos-exp/f-8 10 eos_dash_exp f_dash_8
| x | y | z | |
|---|---|---|---|
| A0 | -1.92969 | 0.0 | -1.92969 |
| A1 | 0.0 | -1.92969 | 1.92969 |
| A2 | -3.43932 | 3.43932 | 3.43932 |
| species | x | y | z | fx | fy | fz | free x | free y | free z |
|---|---|---|---|---|---|---|---|---|---|
| Al | -3.71109103713 | 1.74742547037 | 1.75920312374 | -0.003371 | -0.008528 | 0.015983 | T | T | T |
| Al | -1.75760409593 | -0.206692341706 | 1.74515790595 | 0.004847 | -0.014457 | -0.026033 | T | T | T |
| Ni | -0.0516348669472 | 0.011436871336 | 0.0318889703136 | -0.001476 | 0.022985 | 0.01005 | T | T | T |
| Parameter | Value |
|---|---|
| nbands | 25 |
| encut | 375.0 |
| sigma | 0.1 |
| prec | Normal |
| kpts | [12, 12, 5] |
| xc | PBE |
| txt | - |
| gamma | True |
| Ni | potpawPBE/Ni/POTCAR | 6c814bc7144b98435771aded093fb4ebe527acd9 |
| Al | potpawPBE/Al/POTCAR | c8d9ecb0b6ebec0256c5f5072cee4de6a046dac2 |
1.16 11/eos-exp/f+0 11 eos_dash_exp f_plus_0
| x | y | z | |
|---|---|---|---|
| A0 | -1.83341 | 0.0 | -1.83341 |
| A1 | 0.0 | -1.83341 | 1.83341 |
| A2 | -3.44161 | 3.44161 | 3.44161 |
| species | x | y | z | fx | fy | fz | free x | free y | free z |
|---|---|---|---|---|---|---|---|---|---|
| Al | -1.75834 | -0.07507 | 1.75834 | 0.0 | 0.0 | 0.0 | T | T | T |
| Fe | -3.39510205563 | 3.39510205563 | 3.39510205563 | 0.065417 | -0.065417 | -0.065417 | T | T | T |
| Fe | -3.56318794437 | 1.72977794437 | 1.72977794437 | -0.065417 | 0.065417 | 0.065417 | T | T | T |
| Parameter | Value |
|---|---|
| nbands | 21 |
| encut | 375.0 |
| sigma | 0.1 |
| prec | Normal |
| kpts | [12, 12, 5] |
| xc | PBE |
| txt | - |
| gamma | True |
| Al | potpawPBE/Al/POTCAR | c8d9ecb0b6ebec0256c5f5072cee4de6a046dac2 |
| Fe | potpawPBE/Fe/POTCAR | 201d71adcb39575fc20d69f0abd331ae9c01ed2e |
1.17 11/eos-exp/f+10 11 eos_dash_exp f_plus_10
| x | y | z | |
|---|---|---|---|
| A0 | -1.89259 | 0.0 | -1.89259 |
| A1 | 0.0 | -1.89259 | 1.89259 |
| A2 | -3.55271 | 3.55271 | 3.55271 |
| species | x | y | z | fx | fy | fz | free x | free y | free z |
|---|---|---|---|---|---|---|---|---|---|
| Al | -1.71768562473 | -0.12420477333 | 1.72897811362 | -0.027763 | -0.002052 | 0.049582 | T | T | T |
| Fe | -5.29679962665 | 1.54076691889 | 3.42444208823 | 0.067989 | -0.02176 | -0.05605 | T | T | T |
| Fe | -3.59473474862 | 1.74065785444 | 1.72979979816 | -0.040226 | 0.023812 | 0.006467 | T | T | T |
| Parameter | Value |
|---|---|
| nbands | 21 |
| encut | 375.0 |
| sigma | 0.1 |
| prec | Normal |
| kpts | [12, 12, 5] |
| xc | PBE |
| txt | - |
| gamma | True |
| Al | potpawPBE/Al/POTCAR | c8d9ecb0b6ebec0256c5f5072cee4de6a046dac2 |
| Fe | potpawPBE/Fe/POTCAR | 201d71adcb39575fc20d69f0abd331ae9c01ed2e |
1.18 11/eos-exp/f+12 11 eos_dash_exp f_plus_12
| x | y | z | |
|---|---|---|---|
| A0 | -1.90399 | 0.0 | -1.90399 |
| A1 | 0.0 | -1.90399 | 1.90399 |
| A2 | -3.57411 | 3.57411 | 3.57411 |
| species | x | y | z | fx | fy | fz | free x | free y | free z |
|---|---|---|---|---|---|---|---|---|---|
| Al | -1.71395551429 | -0.137649234072 | 1.73446168186 | 0.003858 | -0.016816 | 0.003362 | T | T | T |
| Fe | -5.30314000606 | 1.54316547516 | 3.42737702355 | 0.034226 | -0.051683 | -0.060287 | T | T | T |
| Fe | -3.60352447965 | 1.74030375892 | 1.7213812946 | -0.038084 | 0.068499 | 0.056924 | T | T | T |
| Parameter | Value |
|---|---|
| nbands | 21 |
| encut | 375.0 |
| sigma | 0.1 |
| prec | Normal |
| kpts | [12, 12, 5] |
| xc | PBE |
| txt | - |
| gamma | True |
| Al | potpawPBE/Al/POTCAR | c8d9ecb0b6ebec0256c5f5072cee4de6a046dac2 |
| Fe | potpawPBE/Fe/POTCAR | 201d71adcb39575fc20d69f0abd331ae9c01ed2e |
1.19 11/eos-exp/f+15 11 eos_dash_exp f_plus_15
| x | y | z | |
|---|---|---|---|
| A0 | -1.92084 | 0.0 | -1.92084 |
| A1 | 0.0 | -1.92084 | 1.92084 |
| A2 | -3.60574 | 3.60574 | 3.60574 |
| species | x | y | z | fx | fy | fz | free x | free y | free z |
|---|---|---|---|---|---|---|---|---|---|
| Al | -1.70274811209 | -0.152372217551 | 1.72799358203 | -0.001016 | -0.015145 | 0.009612 | T | T | T |
| Fe | -5.31888643404 | 1.53641622409 | 3.43283036647 | 0.011105 | -0.02964 | -0.037354 | T | T | T |
| Fe | -3.61583545387 | 1.74492599346 | 1.7223960515 | -0.010088 | 0.044785 | 0.027742 | T | T | T |
| Parameter | Value |
|---|---|
| nbands | 21 |
| encut | 375.0 |
| sigma | 0.1 |
| prec | Normal |
| kpts | [12, 12, 5] |
| xc | PBE |
| txt | - |
| gamma | True |
| Al | potpawPBE/Al/POTCAR | c8d9ecb0b6ebec0256c5f5072cee4de6a046dac2 |
| Fe | potpawPBE/Fe/POTCAR | 201d71adcb39575fc20d69f0abd331ae9c01ed2e |
1.20 11/eos-exp/f+2 11 eos_dash_exp f_plus_2
| x | y | z | |
|---|---|---|---|
| A0 | -1.84555 | 0.0 | -1.84555 |
| A1 | 0.0 | -1.84555 | 1.84555 |
| A2 | -3.4644 | 3.4644 | 3.4644 |
| species | x | y | z | fx | fy | fz | free x | free y | free z |
|---|---|---|---|---|---|---|---|---|---|
| Al | -1.75029374339 | -0.0841326953171 | 1.75144674035 | -0.006952 | -0.003547 | 0.01682 | T | T | T |
| Fe | -5.24418323089 | 1.55674197758 | 3.40127583308 | 0.081175 | -0.052085 | -0.064081 | T | T | T |
| Fe | -3.56770302572 | 1.73165071774 | 1.73049742657 | -0.074224 | 0.055633 | 0.047261 | T | T | T |
| Parameter | Value |
|---|---|
| nbands | 21 |
| encut | 375.0 |
| sigma | 0.1 |
| prec | Normal |
| kpts | [12, 12, 5] |
| xc | PBE |
| txt | - |
| gamma | True |
| Al | potpawPBE/Al/POTCAR | c8d9ecb0b6ebec0256c5f5072cee4de6a046dac2 |
| Fe | potpawPBE/Fe/POTCAR | 201d71adcb39575fc20d69f0abd331ae9c01ed2e |
1.21 11/eos-exp/f+4 11 eos_dash_exp f_plus_4
| x | y | z | |
|---|---|---|---|
| A0 | -1.85754 | 0.0 | -1.85754 |
| A1 | 0.0 | -1.85754 | 1.85754 |
| A2 | -3.4869 | 3.4869 | 3.4869 |
| species | x | y | z | fx | fy | fz | free x | free y | free z |
|---|---|---|---|---|---|---|---|---|---|
| Al | -1.74219271636 | -0.0933112216124 | 1.74481701196 | -0.01609 | -0.003222 | 0.033588 | T | T | T |
| Fe | -5.26036201118 | 1.55320104576 | 3.40867525476 | 0.120223 | -0.071335 | -0.093285 | T | T | T |
| Fe | -3.57161527246 | 1.73238017585 | 1.72972773328 | -0.104133 | 0.074557 | 0.059697 | T | T | T |
| Parameter | Value |
|---|---|
| nbands | 21 |
| encut | 375.0 |
| sigma | 0.1 |
| prec | Normal |
| kpts | [12, 12, 5] |
| xc | PBE |
| txt | - |
| gamma | True |
| Al | potpawPBE/Al/POTCAR | c8d9ecb0b6ebec0256c5f5072cee4de6a046dac2 |
| Fe | potpawPBE/Fe/POTCAR | 201d71adcb39575fc20d69f0abd331ae9c01ed2e |
1.22 11/eos-exp/f+6 11 eos_dash_exp f_plus_6
| x | y | z | |
|---|---|---|---|
| A0 | -1.86937 | 0.0 | -1.86937 |
| A1 | 0.0 | -1.86937 | 1.86937 |
| A2 | -3.50911 | 3.50911 | 3.50911 |
| species | x | y | z | fx | fy | fz | free x | free y | free z |
|---|---|---|---|---|---|---|---|---|---|
| Al | -1.73452887124 | -0.101697405678 | 1.73856189969 | -0.015386 | -0.012456 | 0.039644 | T | T | T |
| Fe | -5.27502082144 | 1.54772132046 | 3.4142939235 | 0.127115 | -0.055701 | -0.087484 | T | T | T |
| Fe | -3.57645030732 | 1.73441608522 | 1.73036417681 | -0.111729 | 0.068157 | 0.04784 | T | T | T |
| Parameter | Value |
|---|---|
| nbands | 21 |
| encut | 375.0 |
| sigma | 0.1 |
| prec | Normal |
| kpts | [12, 12, 5] |
| xc | PBE |
| txt | - |
| gamma | True |
| Al | potpawPBE/Al/POTCAR | c8d9ecb0b6ebec0256c5f5072cee4de6a046dac2 |
| Fe | potpawPBE/Fe/POTCAR | 201d71adcb39575fc20d69f0abd331ae9c01ed2e |
1.23 11/eos-exp/f+8 11 eos_dash_exp f_plus_8
| x | y | z | |
|---|---|---|---|
| A0 | -1.88105 | 0.0 | -1.88105 |
| A1 | 0.0 | -1.88105 | 1.88105 |
| A2 | -3.53104 | 3.53104 | 3.53104 |
| species | x | y | z | fx | fy | fz | free x | free y | free z |
|---|---|---|---|---|---|---|---|---|---|
| Al | -1.72649034784 | -0.11573354282 | 1.73674863604 | -0.016594 | -0.002774 | 0.032443 | T | T | T |
| Fe | -5.28013390733 | 1.54597101428 | 3.41783536417 | 0.029827 | -0.01744 | -0.039596 | T | T | T |
| Fe | -3.59105574483 | 1.73852252854 | 1.7286359998 | -0.013233 | 0.020213 | 0.007153 | T | T | T |
| Parameter | Value |
|---|---|
| nbands | 21 |
| encut | 375.0 |
| sigma | 0.1 |
| prec | Normal |
| kpts | [12, 12, 5] |
| xc | PBE |
| txt | - |
| gamma | True |
| Al | potpawPBE/Al/POTCAR | c8d9ecb0b6ebec0256c5f5072cee4de6a046dac2 |
| Fe | potpawPBE/Fe/POTCAR | 201d71adcb39575fc20d69f0abd331ae9c01ed2e |
1.24 11/eos-exp/f-10 11 eos_dash_exp f_dash_10
| x | y | z | |
|---|---|---|---|
| A0 | -1.77014 | 0.0 | -1.77014 |
| A1 | 0.0 | -1.77014 | 1.77014 |
| A2 | -3.32284 | 3.32284 | 3.32284 |
| species | x | y | z | fx | fy | fz | free x | free y | free z |
|---|---|---|---|---|---|---|---|---|---|
| Al | -1.7975784663 | -0.01829295971 | 1.78049380651 | -0.007299 | 0.023676 | -0.013472 | T | T | T |
| Fe | -0.0656240800248 | 0.0224466618076 | 0.0395349460384 | 0.084793 | -0.065991 | -0.04627 | T | T | T |
| Fe | -3.53058745368 | 1.7228162979 | 1.74035124745 | -0.077494 | 0.042315 | 0.059741 | T | T | T |
| Parameter | Value |
|---|---|
| nbands | 21 |
| encut | 375.0 |
| sigma | 0.1 |
| prec | Normal |
| kpts | [12, 12, 5] |
| xc | PBE |
| txt | - |
| gamma | True |
| Al | potpawPBE/Al/POTCAR | c8d9ecb0b6ebec0256c5f5072cee4de6a046dac2 |
| Fe | potpawPBE/Fe/POTCAR | 201d71adcb39575fc20d69f0abd331ae9c01ed2e |
1.25 11/eos-exp/f-12 11 eos_dash_exp f_dash_12
| x | y | z | |
|---|---|---|---|
| A0 | -1.75693 | 0.0 | -1.75693 |
| A1 | 0.0 | -1.75693 | 1.75693 |
| A2 | -3.29804 | 3.29804 | 3.29804 |
| species | x | y | z | fx | fy | fz | free x | free y | free z |
|---|---|---|---|---|---|---|---|---|---|
| Al | -1.80588201814 | -0.00576933899531 | 1.78487378934 | -0.004797 | 0.023557 | -0.019673 | T | T | T |
| Fe | -0.0889524804644 | 0.0361683086543 | 0.057258635695 | 0.086292 | -0.05927 | -0.035757 | T | T | T |
| Fe | -3.52375550139 | 1.72137103034 | 1.74304757497 | -0.081495 | 0.035713 | 0.05543 | T | T | T |
| Parameter | Value |
|---|---|
| nbands | 21 |
| encut | 375.0 |
| sigma | 0.1 |
| prec | Normal |
| kpts | [12, 12, 5] |
| xc | PBE |
| txt | - |
| gamma | True |
| Al | potpawPBE/Al/POTCAR | c8d9ecb0b6ebec0256c5f5072cee4de6a046dac2 |
| Fe | potpawPBE/Fe/POTCAR | 201d71adcb39575fc20d69f0abd331ae9c01ed2e |
1.26 11/eos-exp/f-15 11 eos_dash_exp f_dash_15
| x | y | z | |
|---|---|---|---|
| A0 | -1.73673 | 0.0 | -1.73673 |
| A1 | 0.0 | -1.73673 | 1.73673 |
| A2 | -3.26013 | 3.26013 | 3.26013 |
| species | x | y | z | fx | fy | fz | free x | free y | free z |
|---|---|---|---|---|---|---|---|---|---|
| Al | -1.81850401219 | 0.0184814979188 | 1.7846748897 | -0.013651 | 0.006945 | 0.022302 | T | T | T |
| Fe | -0.121256908412 | 0.0564193466513 | 0.0901670119937 | 0.073536 | -0.071854 | -0.082079 | T | T | T |
| Fe | -3.5167390794 | 1.71477915543 | 1.7482480983 | -0.059885 | 0.064909 | 0.059777 | T | T | T |
| Parameter | Value |
|---|---|
| nbands | 21 |
| encut | 375.0 |
| sigma | 0.1 |
| prec | Normal |
| kpts | [12, 12, 5] |
| xc | PBE |
| txt | - |
| gamma | True |
| Al | potpawPBE/Al/POTCAR | c8d9ecb0b6ebec0256c5f5072cee4de6a046dac2 |
| Fe | potpawPBE/Fe/POTCAR | 201d71adcb39575fc20d69f0abd331ae9c01ed2e |
1.27 11/eos-exp/f-2 11 eos_dash_exp f_dash_2
| x | y | z | |
|---|---|---|---|
| A0 | -1.8211 | 0.0 | -1.8211 |
| A1 | 0.0 | -1.8211 | 1.8211 |
| A2 | -3.41851 | 3.41851 | 3.41851 |
| species | x | y | z | fx | fy | fz | free x | free y | free z |
|---|---|---|---|---|---|---|---|---|---|
| Al | -1.76538350661 | -0.067067453209 | 1.76453861606 | -0.008756 | 0.02094 | -0.003441 | T | T | T |
| Fe | -3.39061275864 | 3.3875708672 | 3.38853117003 | 0.024749 | -0.062564 | -0.052235 | T | T | T |
| Fe | -3.56063373475 | 1.72930658601 | 1.73015021391 | -0.015993 | 0.041624 | 0.055675 | T | T | T |
| Parameter | Value |
|---|---|
| nbands | 21 |
| encut | 375.0 |
| sigma | 0.1 |
| prec | Normal |
| kpts | [12, 12, 5] |
| xc | PBE |
| txt | - |
| gamma | True |
| Al | potpawPBE/Al/POTCAR | c8d9ecb0b6ebec0256c5f5072cee4de6a046dac2 |
| Fe | potpawPBE/Fe/POTCAR | 201d71adcb39575fc20d69f0abd331ae9c01ed2e |
1.28 11/eos-exp/f-4 11 eos_dash_exp f_dash_4
| x | y | z | |
|---|---|---|---|
| A0 | -1.80863 | 0.0 | -1.80863 |
| A1 | 0.0 | -1.80863 | 1.80863 |
| A2 | -3.3951 | 3.3951 | 3.3951 |
| species | x | y | z | fx | fy | fz | free x | free y | free z |
|---|---|---|---|---|---|---|---|---|---|
| Al | -1.77477734473 | -0.0548385641908 | 1.77147863672 | 0.018222 | -8.4e-05 | -0.038433 | T | T | T |
| Fe | -3.38991272702 | 3.37906159113 | 3.38279256816 | 0.054526 | -0.094194 | -0.078195 | T | T | T |
| Fe | -3.55193992824 | 1.72558697306 | 1.72894879512 | -0.072747 | 0.094278 | 0.116628 | T | T | T |
| Parameter | Value |
|---|---|
| nbands | 21 |
| encut | 375.0 |
| sigma | 0.1 |
| prec | Normal |
| kpts | [12, 12, 5] |
| xc | PBE |
| txt | - |
| gamma | True |
| Al | potpawPBE/Al/POTCAR | c8d9ecb0b6ebec0256c5f5072cee4de6a046dac2 |
| Fe | potpawPBE/Fe/POTCAR | 201d71adcb39575fc20d69f0abd331ae9c01ed2e |
1.29 11/eos-exp/f-6 11 eos_dash_exp f_dash_6
| x | y | z | |
|---|---|---|---|
| A0 | -1.79598 | 0.0 | -1.79598 |
| A1 | 0.0 | -1.79598 | 1.79598 |
| A2 | -3.37135 | 3.37135 | 3.37135 |
| species | x | y | z | fx | fy | fz | free x | free y | free z |
|---|---|---|---|---|---|---|---|---|---|
| Al | -1.78297699606 | -0.0446680285269 | 1.77821103953 | 0.028987 | -0.000194 | -0.062433 | T | T | T |
| Fe | -0.0156774252598 | -0.000431572871362 | 0.00517398273374 | 0.050099 | -0.112472 | -0.086264 | T | T | T |
| Fe | -3.54662557868 | 1.7235596014 | 1.72848497774 | -0.079085 | 0.112666 | 0.148697 | T | T | T |
| Parameter | Value |
|---|---|
| nbands | 21 |
| encut | 375.0 |
| sigma | 0.1 |
| prec | Normal |
| kpts | [12, 12, 5] |
| xc | PBE |
| txt | - |
| gamma | True |
| Al | potpawPBE/Al/POTCAR | c8d9ecb0b6ebec0256c5f5072cee4de6a046dac2 |
| Fe | potpawPBE/Fe/POTCAR | 201d71adcb39575fc20d69f0abd331ae9c01ed2e |
1.30 11/eos-exp/f-8 11 eos_dash_exp f_dash_8
| x | y | z | |
|---|---|---|---|
| A0 | -1.78315 | 0.0 | -1.78315 |
| A1 | 0.0 | -1.78315 | 1.78315 |
| A2 | -3.34727 | 3.34727 | 3.34727 |
| species | x | y | z | fx | fy | fz | free x | free y | free z |
|---|---|---|---|---|---|---|---|---|---|
| Al | -1.78874833405 | -0.029531043967 | 1.77374404304 | -0.022672 | 0.030355 | 0.013661 | T | T | T |
| Fe | -0.0461476403345 | 0.00738880489013 | 0.0220586228803 | 0.121535 | -0.072896 | -0.06196 | T | T | T |
| Fe | -3.53446402561 | 1.72468223908 | 1.74014733408 | -0.098863 | 0.042541 | 0.048299 | T | T | T |
| Parameter | Value |
|---|---|
| nbands | 21 |
| encut | 375.0 |
| sigma | 0.1 |
| prec | Normal |
| kpts | [12, 12, 5] |
| xc | PBE |
| txt | - |
| gamma | True |
| Al | potpawPBE/Al/POTCAR | c8d9ecb0b6ebec0256c5f5072cee4de6a046dac2 |
| Fe | potpawPBE/Fe/POTCAR | 201d71adcb39575fc20d69f0abd331ae9c01ed2e |
2 Tabulating the results in column mode
We can use column view to see the results in a tabular like form. For example, here we capture the VOLUME and TOTALENERGY properties.
| ITEM | VOLUME | TOTALENERGY |
|---|---|---|
| * Using org-entries like a database | ||
| ** Generating the entries | ||
| * 10/eos-exp/f+0 :10:eosdashexp:fplus0: | 41.7621235495 | -13.997605 |
| * 10/eos-exp/f+10 :10:eosdashexp:fplus10: | 45.9384934449 | -13.874719 |
| * 10/eos-exp/f+12 :10:eosdashexp:fplus12: | 46.7733446838 | -13.826981 |
| * 10/eos-exp/f+15 :10:eosdashexp:fplus15: | 48.0264189364 | -13.744565 |
| * 10/eos-exp/f+2 :10:eosdashexp:fplus2: | 42.5973719405 | -13.991505 |
| * 10/eos-exp/f+4 :10:eosdashexp:fplus4: | 43.4325784041 | -13.97507 |
| * 10/eos-exp/f+6 :10:eosdashexp:fplus6: | 44.2677528152 | -13.949606 |
| * 10/eos-exp/f+8 :10:eosdashexp:fplus8: | 45.10318841 | -13.915655 |
| * 10/eos-exp/f-10 :10:eosdashexp:fdash10: | 37.5859624695 | -13.819891 |
| * 10/eos-exp/f-12 :10:eosdashexp:fdash12: | 36.75057064 | -13.730761 |
| * 10/eos-exp/f-15 :10:eosdashexp:fdash15: | 35.4979860465 | -13.553386 |
| * 10/eos-exp/f-2 :10:eosdashexp:fdash2: | 40.9267763602 | -13.991485 |
| * 10/eos-exp/f-4 :10:eosdashexp:fdash4: | 40.0914758244 | -13.971902 |
| * 10/eos-exp/f-6 :10:eosdashexp:fdash6: | 39.2564933754 | -13.938219 |
| * 10/eos-exp/f-8 :10:eosdashexp:fdash8: | 38.4210237246 | -13.888471 |
| * 11/eos-exp/f+0 :11:eosdashexp:fplus0: | 34.7058033185 | -20.281953 |
| * 11/eos-exp/f+10 :11:eosdashexp:fplus10: | 38.1763228931 | -20.095119 |
| * 11/eos-exp/f+12 :11:eosdashexp:fplus12: | 38.870353968 | -20.024412 |
| * 11/eos-exp/f+15 :11:eosdashexp:fplus15: | 39.9114994655 | -19.90227 |
| * 11/eos-exp/f+2 :11:eosdashexp:fplus2: | 35.3998087733 | -20.271762 |
| * 11/eos-exp/f+4 :11:eosdashexp:fplus4: | 36.0941730661 | -20.24583 |
| * 11/eos-exp/f+6 :11:eosdashexp:fplus6: | 36.7882199604 | -20.206804 |
| * 11/eos-exp/f+8 :11:eosdashexp:fplus8: | 37.4821566447 | -20.157092 |
| * 11/eos-exp/f-10 :11:eosdashexp:fdash10: | 31.2353169019 | -20.038179 |
| * 11/eos-exp/f-12 :11:eosdashexp:fdash12: | 30.5411995447 | -19.912894 |
| * 11/eos-exp/f-15 :11:eosdashexp:fdash15: | 29.4999164187 | -19.662755 |
| * 11/eos-exp/f-2 :11:eosdashexp:fdash2: | 34.0114931233 | -20.275411 |
| * 11/eos-exp/f-4 :11:eosdashexp:fdash4: | 33.31756747 | -20.249632 |
| * 11/eos-exp/f-6 :11:eosdashexp:fdash6: | 32.6233149155 | -20.202731 |
| * 11/eos-exp/f-8 :11:eosdashexp:fdash8: | 31.9291793012 | -20.133749 |
| ** Tabulating the results in column mode | ||
| ** org-map-entries to get the data | ||
| ** Getting specific data from a calculation | ||
| ** Summary |
That is more or less what we want, but we cannot use that table as a data source very directly. While you can use it as a data source, you just get a string of the whole table, e.g.
print data.__class__
<type 'str'>
You could parse that string, or try converting it to a table, but we skip that for now. In the next section we consider another approach that more directly gets at the data.
3 org-map-entries to get the data
Let us try a different approach. Here we map over headlines that match a criteria, and get specific data. We add a header of labels and horizontal line to the beginning of the table. Here we find the results tagged with 11, and that match fdash* (we actually use a regular expression).
(cons '(label volume energy)
(cons 'hline
(org-map-entries
(lambda ()
(list (nth 4 (org-heading-components))
(org-entry-get (point) "VOLUME")
(org-entry-get (point) "TOTAL_ENERGY")))
;; this next line specifies headlines tagged with 11
;; and that match the regular expression in {}
"+11+{f_dash_.*}")))
| label | volume | energy |
|---|---|---|
| 11/eos-exp/f-10 | 31.2353169019 | -20.038179 |
| 11/eos-exp/f-12 | 30.5411995447 | -19.912894 |
| 11/eos-exp/f-15 | 29.4999164187 | -19.662755 |
| 11/eos-exp/f-2 | 34.0114931233 | -20.275411 |
| 11/eos-exp/f-4 | 33.31756747 | -20.249632 |
| 11/eos-exp/f-6 | 32.6233149155 | -20.202731 |
| 11/eos-exp/f-8 | 31.9291793012 | -20.133749 |
Now, since there are strings in the table (or in the results from the named code-block), we will have to cast the results to floats using a numpy array. Note it appears that we actually run the code block to get the results, they do not come from the table. The table is just helpful to know what data you are using.
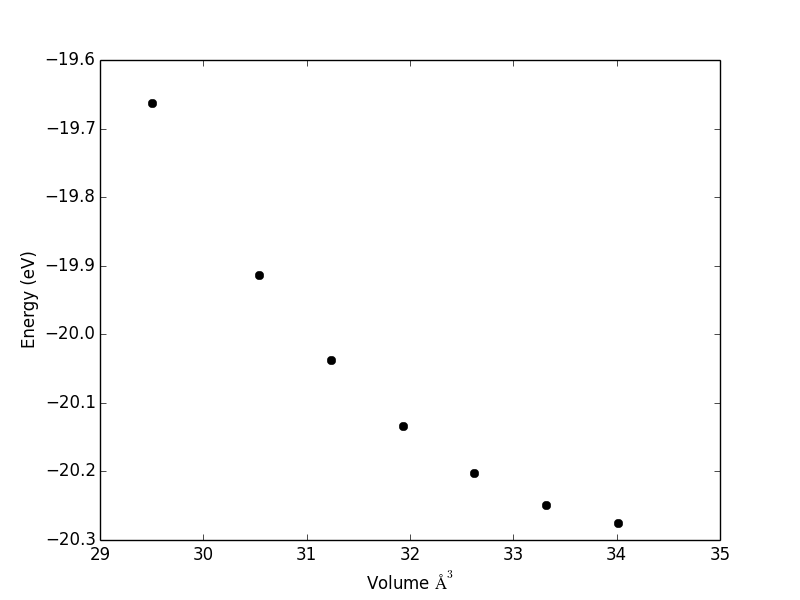
import matplotlib.pyplot as plt import numpy as np print E plt.plot(np.array(V), np.array(E), 'ko') plt.xlabel('Volume $\AA^3$') plt.ylabel('Energy (eV)') plt.savefig('11-eos.png')
['-20.038179', '-19.912894', '-19.662755', '-20.275411', '-20.249632', '-20.202731', '-20.133749']
And here you have the figure. We extracted some data into an org-file in human readable form. Then, we extracted the data from those entries to a table, and finally plotted it.
4 Getting specific data from a calculation
Say you want the stress from some calculation. Each heading is identified by a CUSTOMID, so we can temporarily go to that headline, and get the property. Easy!
(save-excursion (org-open-link-from-string "[[#10/eos-exp/f+0]]") (org-entry-get (point) "STRESS"))
[-0.003191325893994518, -0.003191325893994518, -0.003191325893994518, -0.00042576435655845916, 0.00042576435655845916, 0.00042576435655845916]
Note, however, that the value returned is a string, so we need to convert it to numbers if we want to do anything with it, e.g. find the maximum component. Here, we eval the string in Python to make it a list, and then find the maximum value.
import numpy as np print np.abs(np.array(eval(stress))).max()
0.00319132589399
Next, we get the unit cell from that calculation, so we can calculate the volume using python. The unit cell is stored in a table and we extract the unit cell vectors from it in the code block header.
import numpy as np print('cell = {}'.format(np.array(data))) print('Volume = {}'.format(np.linalg.det(np.array(data))))
cell = [[-1.98408 0. -1.98408] [ 0. -1.98408 1.98408] [-3.53625 3.53625 3.53625]] Volume = 41.7621235495
Compare that to the volume we computed when the entry was generated, and you can see they are the same.
(save-excursion (org-open-link-from-string "[[#10/eos-exp/f+0]]") (org-entry-get (point) "VOLUME"))
41.7621235495
The last example is getting the formula for the calculation. The atoms positions are stored in a table in the entry, and the species for each atom is stored in the first column. We extract the species into a variable, then then get counts for each species, and finally construct a string representation of the formula.
(let* ((unique-species '()) (counts)) ;; first get unique species (mapcar (lambda (s) (add-to-list 'unique-species s)) species) ;; now get counts (setq counts (mapcar (lambda (s) (--count (string= it s) species)) unique-species)) ;; Now put them together (mapconcat 'identity (cl-mapcar (lambda (s n) (format "%s%s" s n)) unique-species counts) ""))
Ni1Al2
Suppose we want to sort the elements alphabetically. We can make a list of cons cells of element and count, sort the list by element, and then put them back together. We do this in lisp.
(let* ((unique-species '()) (counts)) ;; first get unique species (mapcar (lambda (s) (add-to-list 'unique-species s)) species) ;; now get counts (setq counts (mapcar (lambda (s) (--count (string= it s) species)) unique-species)) (mapconcat (lambda (x) (format "%s%s" (car x) (cdr x))) (cl-sort (cl-mapcar (lambda (a b) (cons a b)) unique-species counts) 'string-lessp :key 'car) ""))
Al2Ni1
That is not too bad. It is a little verbose, some of that could be put into a library to be reused. For example, here is the same code in Python for the unsorted formula
from collections import Counter counts = Counter(species) print ''.join(['{0}{1}'.format(x, counts[x]) for x in counts])
Ni1Al2
and for the sorted formula.
from collections import Counter counts = Counter(species) print ''.join(['{0}{1}'.format(x, counts[x]) for x in sorted(counts.keys())])
Al2Ni1
Obviously a lot must be happening behind the scenes in those short, dense python blocks. For example, the list comprehension syntax in Python is functionally similar to the mapcar code, and the join function is similar to the mapconcat code in lisp.
5 Summary
This has some potential. It is a hybrid approach to human and machine readable data, that enables inline analysis. You can mix programming languages, which is great if one language has unique capabilities or is just more convenient. There are some limitations, mostly that org-properties are read as strings, and they have to be converted to the right data type, e.g. numbers or lists or arrays, etc… But, these can be saved as forms that can be directly evaled in some languages. On the other hand, it is possible to do analysis on another machine where the original data does not exist.
This is a very different mental model of data than I am used to. We typically have thought about data in files, and paths to get to that data. Here, data is in headings, and we use links to get to the data. We also use different syntax to link data to code blocks. That means in some cases the code blocks cannot be run independently of this file.
Copyright (C) 2014 by John Kitchin. See the License for information about copying.
Org-mode version = 8.2.7c