Serializing jasp calculations as json data
Posted October 19, 2013 at 02:33 PM | categories: jasp, ase, vasp | tags:
Updated October 19, 2013 at 03:10 PM
Table of Contents
We use VASPto calculate materials properties in our research We use the jasppython module we have developed to setup, run and analyze those calculations. One of the things we have worked on developing recently is to more transparently share how do this kind of work by using org-mode supporting information files. Doing this should make our research more reproducible, and allow others to build off of it more easily.
We have run into the following problem trying to share VASP results however. The VASP license prohibits us from sharing the POTCAR files that are used to run the calculations. That is unfortunate, but since these files are also what give VASP some competitive advantage, they are protected, and we agreed to that when we bought the license. The problem is that the jasp module requires the POTCAR files to work, so without them, our scripts are not reproducible by researchers without a VASP license.
So, we have been looking at new ways to share the data from our calculations. In this post, we consider representing the calculation as a JSON file. We will look at a couple of new features built into the development branch of jasp
1 The simplest case of a simple calculation
Here we setup and run a simple calculation, and output the JSON file.
from ase import Atoms, Atom from jasp import * import numpy as np np.set_printoptions(precision=3, suppress=True) co = Atoms([Atom('C',[0, 0, 0]), Atom('O',[1.2, 0, 0])], cell=(6., 6., 6.)) with jasp('molecules/simple-co', #output dir xc='PBE', # the exchange-correlation functional nbands=6, # number of bands encut=350, # planewave cutoff ismear=1, # Methfessel-Paxton smearing sigma=0.01,# very small smearing factor for a molecule atoms=co) as calc: print 'energy = {0} eV'.format(co.get_potential_energy()) print co.get_forces() with open('JSON', 'w') as f: f.write(calc.json)
energy = -14.687906 eV [[ 5.095 0. 0. ] [-5.095 0. 0. ]]
Now, we can analyze the JSON file independently of jasp. The json data contains all the inputs we used for the VASP calculation, the atomic geometry, and many of the outputs of the calculation. Here is the JSONfile.
import json with open('molecules/simple-co/JSON', 'rb') as f: d = json.loads(f.read()) print('The energy is {0}'.format(d['data']['total_energy'])) print('The forces are {0}'.format(d['data']['forces']))
The energy is -14.687906 The forces are [[5.095488, 0.0, 0.0], [-5.095488, 0.0, 0.0]]
2 Including extra information in the JSON file
If we use a slightly different syntax, we can also include the total DOS in the JSON file.
from jasp import * with jasp('molecules/simple-co') as calc: with open('JSON-DOS', 'w') as f: f.write(calc_to_json(calc, dos=True))
To illustrate that we have done that, let us plot the DOS without using jasp from the JSON-DOSfile.
import json import matplotlib.pyplot as plt with open('molecules/simple-co/JSON-DOS', 'rb') as f: d = json.loads(f.read()) energies = d['data']['dos']['e'] dos = d['data']['dos']['dos'] plt.plot(energies, dos) plt.savefig('molecules/simple-co/dos.png')
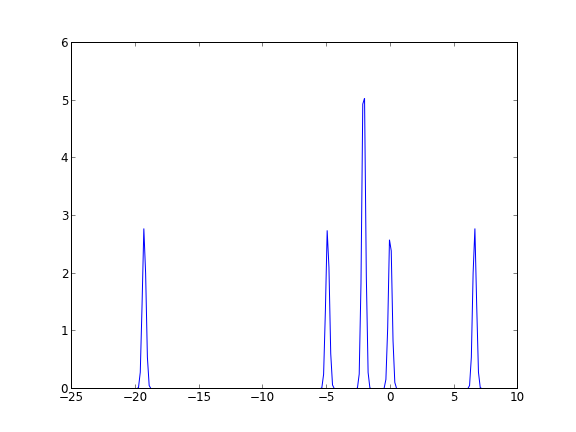
We are still working on getting atom-projected DOS into the json file, and ensuring that all the spin cases are handled (e.g. the spin-up and spin-down DOS).
3 Limitations?
JSON is flexible, and can store text and numeric data. It does not store numpy arrays, but rather it is limited to storing lists of data. You would have to convert them back to arrays if you want to do array math. You probably wouldn't want to store a 3d array of electron density in this format, although it probably isn't worse than a CUBE file format. We haven't tested these files very significantly yet at a large scale to see how fast it is to read from lots of them.
Nonetheless, this looks like a reasonable format to share data in human and machine readable form, without violating the VASP licence conditions.
Copyright (C) 2013 by John Kitchin. See the License for information about copying.