Using Custodian to help converge an optimization problem
Posted September 19, 2023 at 03:34 PM | categories: programming, optimization | tags:
Updated September 19, 2023 at 03:35 PM
In high-throughput calculations, some fraction of them usually fail for some reason. Sometimes it is easy to fix these calculations and re-run them successfully, for example, you might just need a different initialization, or to increase memory or the number of allowed steps, etc. custodian is a tool that is designed for this purpose.
The idea is we make a function to do what we want that has arguments that control that. We need a function that can examine the output of the function and determine if it succeeded, and if it didn't succeed to say what new arguments to try next. Then we run the function in custodian and let it take care of rerunning with new arguments until it either succeeds, or tries too many times.
The goal here is to use custodian to fix a problem optimization. The example is a little contrived, we set a number of iterations artificially low so that the minimization fails by reaching the maximum number of iterations. Custodian will catch this, and increase the number of iterations until it succeeds. Here is the objective function:
import matplotlib.pyplot as plt import numpy as np def objective(x): return np.exp(x**2) - 10*np.exp(x) x = np.linspace(0, 2) plt.plot(x, objective(x)) plt.xlabel('x') plt.ylabel('y');
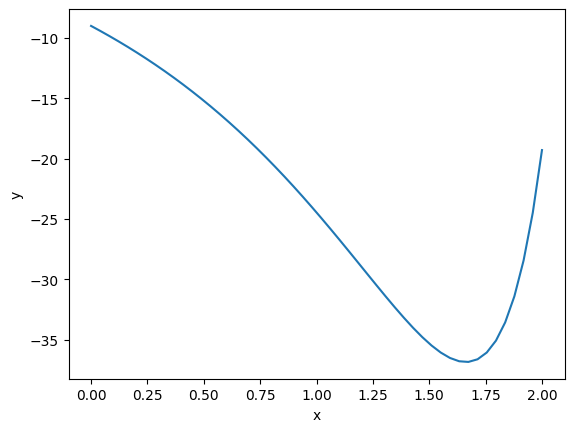
Clearly there is a minimum near 1.75, but with a bad initial guess, and not enough iterations, an optimizer fails here. We can tell it fails from the message here, and the solution is run it again with more iterations.
from scipy.optimize import minimize minimize(objective, 0.0, options={'maxiter': 2})
:RESULTS:
message: Maximum number of iterations has been exceeded.
success: False
status: 1
fun: -36.86289091418059
x: [ 1.661e+00]
nit: 2
jac: [-2.374e-01]
hess_inv: [[ 6.889e-03]]
nfev: 20
njev: 10
:END:
With Custodian you define a "Job". This is a class with params that contain the adjustable arguments in a dictionary, and a run method that stores the results in the params attribute. This is an important step, because the error handlers only get the params, so you need the results in there to inspect them.
The error handlers are another class with a check method that returns True if you should rerun, and a correct method that sets the params to new values to try next. It seems to return some information about what happened. In the correct method, we double the maximum number of iterations allowed, and use the last solution point that failed as the initial guess for the next run.
from custodian.custodian import Custodian, Job, ErrorHandler class Minimizer(Job): def __init__(self, params=None): self.params = params if params else {'maxiter': 2, 'x0': 0} def run(self): sol = minimize(objective, self.params['x0'], options={'maxiter': self.params['maxiter']}) self.params['sol'] = sol class MaximumIterationsExceeded(ErrorHandler): def __init__(self, params): self.params = params def check(self): return self.params['sol'].message == 'Maximum number of iterations has been exceeded.' def correct(self): self.params['maxiter'] *= 2 self.params['x0'] = self.params['sol'].x return {'errors': 'MaximumIterations Exceeded', 'actions': 'maxiter = {self.params["maxiter"]}, x0 = {self.params["x0"]}'}
Now we setup the initial params to try, create a Custodian object with the handler and job, and then run it. The results and final params are stored in the params object.
params = {'maxiter': 1, 'x0': 0} c = Custodian([MaximumIterationsExceeded(params)], [Minimizer(params)], max_errors=5) c.run() for key in params: print(key, params[key])
MaximumIterationsExceeded
MaximumIterationsExceeded
maxiter 4
x0 [1.66250127]
sol message: Optimization terminated successfully.
success: True
status: 0
fun: -36.86307468296398
x: [ 1.662e+00]
nit: 1
jac: [-9.060e-06]
hess_inv: [[1]]
nfev: 6
njev: 3
Note that params is modified, and finally has the maxiter value that worked, and the solution in it. You can see we had to rerun this problem twice before it succeeded, but this happened automatically after the setup. This example is easy because we can simply increase the maxiter value, and no serious logic is needed. Other use cases might include try it again with another solver, try again with a different initial guess, etc.
It feels a little heavyweight to define the classes, and to store the results in params here, but this was overall under an hour of work to put it all together, starting from scratch with the Custodian documentation from the example on the front page. You can do more sophisticated things, including having multiple error handlers. Overall, for a package designed for molecular simulations, this worked well for a different kind of problem.
Copyright (C) 2023 by John Kitchin. See the License for information about copying.
Org-mode version = 9.7-pre